TrvaA.00834.a
macrophage migration inhibitory factor, mif, putative
| CENTER ID: | TrvaA.00834.a |
| ORGANISM: | Trichomonas vaginalis ATCC PRA-98 / G3 |
| ASSOCIATED DISEASE: | |
| CURRENT STATUS: | in PDB |
| COMMUNITY REQUEST: | True |
| NIH RISK GROUP: | unclassified |
| SELECT AGENT: | False |
| NIH PRIORITY pathogens category: |
unclassified |
Ordering Clones & Proteins
If there are materials available for this target, they will be listed below.
Materials can be ordered from SSGCID using the button in the "order material" column.
Clicking the button will add the material to a virtual cart.
You may order multiple materials at a time at no cost to you, as this contract is funded
by NIAID. When you are ready to place your order, click the "Place Order" link which will
appear in the top right corner of the page after you place your first item in your cart.
Proteins
| CENTER REFERENCE ID |
DOMAIN/REGION DESCRIPTION |
INFO | AA START |
AA STOP |
ORDER MATERIAL |
|---|---|---|---|---|---|
| TrvaA.00834.a.UX1.PW39224 | Full length(TrvaA.00834.a) | 1 | 115 |
Structures
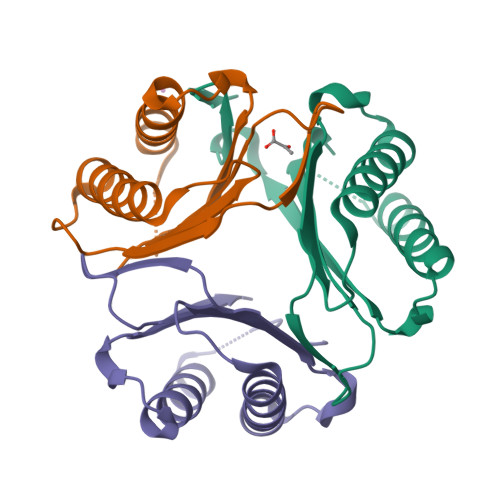
8UR2
DEPOSITED: 10/25/2023
DETERMINATION: XRay
CLONE: TrvaA.00834.a.UX1.GE45037
PROTEIN: TrvaA.00834.a.UX1.PW39224
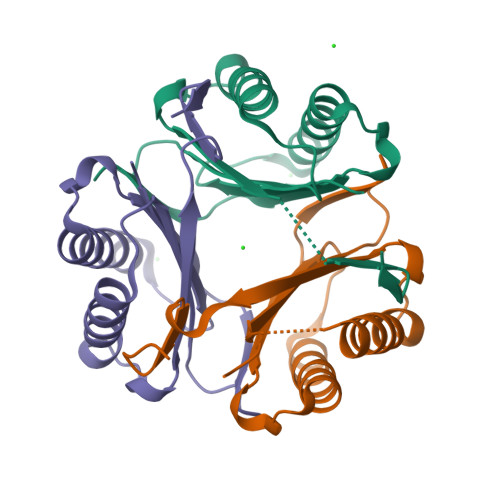
8UR4
DEPOSITED: 10/25/2023
DETERMINATION: XRay
CLONE: TrvaA.00834.a.UX1.GE45037
PROTEIN: TrvaA.00834.a.UX1.PW39224
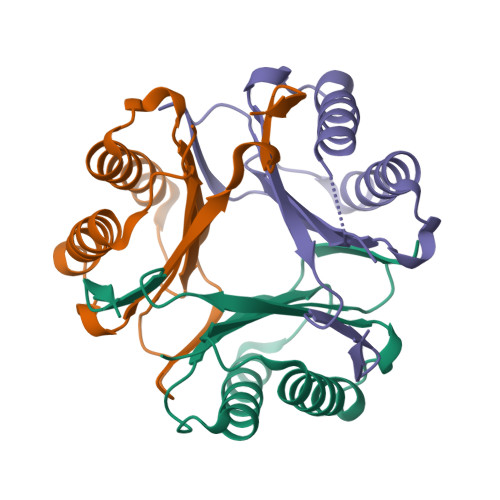
8UZ4
DEPOSITED: 11/14/2023
DETERMINATION: XRay
CLONE: TrvaA.00834.a.UX1.GE45037
PROTEIN: TrvaA.00834.a.UX1.PW39224
Publications by SSGCID
Structures of Trichomonas vaginalis Macrophage Migratory Inhibitory Factor (TvMIF)
Barrett LK, Myler PJ, Subramanian S, Van Voorhis WC, Asojo OA, Battaile KP, Craig J, Lovell S, Liu L, Harmon EK, Dawson OCO, Darwiche R, Srivastava A, Nair A, Gao R
Acta Crystallogr F Struct Biol Commun - 2024
volume 80, issue 12, pages 341-347
PMID: 39601418; PMCID: PMC11614108
External Resources
| RESOURCE | REFERENCE ID |
|---|---|
| EuPathDB: | TrichDB:TVAG_219770 |
| EuPathDB: | TrichDB:TVAGG3_0683450 |
| UniProt: | A2DXT4 |
Sequences
These sequences are the native gene sequence; sequences of constructs derived from these sequences may differ due to codon optimization or other protocols.
To find the specific sequence of any material you may have ordered, click on the "info" button next to the name of that material.
AA Sequence
MPALVIKTNA KFTEEEKSKA TEELGNIVSK VLGKPISYVM VTLEDGVAVR FGGSDEKAAF MSLMSIGGLN RAVNKRASAA LTKWFTDHGF QGDRIYIVFN PKSAEDWGFN GDTFA
NT Sequence
ATGCCTGCTC TTGTTATCAA GACCAACGCT AAATTCACAG AGGAAGAGAA GTCCAAGGCC ACCGAGGAAC TCGGAAACAT TGTTTCCAAG GTTCTTGGAA AGCCAATCTC TTATGTCATG GTCACATTAG AGGATGGTGT TGCTGTTAGA TTCGGTGGCA GTGACGAGAA GGCTGCTTTC ATGTCCCTCA TGTCAATTGG TGGACTTAAC AGAGCTGTTA ACAAGAGGGC ATCTGCTGCT CTCACAAAGT GGTTCACAGA CCATGGTTTC CAGGGCGATC GCATTTACAT CGTCTTCAAT CCAAAGAGTG CTGAGGACTG GGGCTTCAAT GGTGACACAT TTGCC
Details for TrvaA.00834.a.UX1.PW39224
| PURIFICATION DATe: | 8/1/2023 |
| CONCENTRATION: | 35.4mg/ml |
| OBSERVED MW: | 15kDa |
| EXPRESSION LEVEL: | Low Expression |
| PROTEIN PURIFICATION BUFFER: | 25 mM HEPES pH 7.0, 500 mM NaCl, 5% Glycerol, 2 mM DTT, 0.025% Azide |
| EXPRESSION HOST: | BL 21 (DE3) Rosetta |
| VIAL COUNT (approx.): | 63 |
| VIAL VOLUME: | 110µl |
| PERCENT IDENTITY: | data unavailable |
| PERCENT COVERAGE: | data unavailable |
Protocol Notes
notes unavailable
Validated AA Sequence
sequence unavailable
Validated NT Sequence
sequence unavailable
Expressed Protein Sequence
MAHHHHHHMG TLEAQTQGPG SMPALVIKTN AKFTEEEKSK ATEELGNIVS KVLGKPISYV MVTLEDGVAV RFGGSDEKAA FMSLMSIGGL NRAVNKRASA ALTKWFTDHG FQGDRIYIVF NPKSAEDWGF NGDTFA
Full NT Sequence (Expression Vector + Insert)
TGGCGAATGG GACGCGCCCT GTAGCGGCGC ATTAAGCGCG GCGGGTGTGG TGGTTACGCG CAGCGTGACC GCTACACTTG CCAGCGCCCT AGCGCCCGCT CCTTTCGCTT TCTTCCCTTC CTTTCTCGCC ACGTTCGCCG GCTTTCCCCG TCAAGCTCTA AATCGGGGGC TCCCTTTAGG GTTCCGATTT AGTGCTTTAC GGCACCTCGA CCCCAAAAAA CTTGATTAGG GTGATGGTTC ACGTAGTGGG CCATCGCCCT GATAGACGGT TTTTCGCCCT TTGACGTTGG AGTCCACGTT CTTTAATAGT GGACTCTTGT TCCAAACTGG AACAACACTC AACCCTATCT CGGTCTATTC TTTTGATTTA TAAGGGATTT TGCCGATTTC GGCCTATTGG TTAAAAAATG AGCTGATTTA ACAAAAATTT AACGCGAATT TTAACAAAAT ATTAACGCTT ACAATTTAGG TGGCACTTTT CGGGGAAATG TGCGCGGAAC CCCTATTTGT TTATTTTTCT AAATACATTC AAATATGTAT CCGCTCATGA ATTAATTCTT AGAAAAACTC ATCGAGCATC AAATGAAACT GCAATTTATT CATATCAGGA TTATCAATAC CATATTTTTG AAAAAGCCGT TTCTGTAATG AAGGAGAAAA CTCACCGAGG CAGTTCCATA GGATGGCAAG ATCCTGGTAT CGGTCTGCGA TTCCGACTCG TCCAACATCA ATACAACCTA TTAATTTCCC CTCGTCAAAA ATAAGGTTAT CAAGTGAGAA ATCACCATGA GTGACGACTG AATCCGGTGA GAATGGCAAA AGTTTATGCA TTTCTTTCCA GACTTGTTCA ACAGGCCAGC CATTACGCTC GTCATCAAAA TCACTCGCAT CAACCAAACC GTTATTCATT CGTGATTGCG CCTGAGCGAG ACGAAATACG CGATCGCTGT TAAAAGGACA ATTACAAACA GGAATCGAAT GCAACCGGCG CAGGAACACT GCCAGCGCAT CAACAATATT TTCACCTGAA TCAGGATATT CTTCTAATAC CTGGAATGCT GTTTTCCCGG GGATCGCAGT GGTGAGTAAC CATGCATCAT CAGGAGTACG GATAAAATGC TTGATGGTCG GAAGAGGCAT AAATTCCGTC AGCCAGTTTA GTCTGACCAT CTCATCTGTA ACATCATTGG CAACGCTACC TTTGCCATGT TTCAGAAACA ACTCTGGCGC ATCGGGCTTC CCATACAATC GATAGATTGT CGCACCTGAT TGCCCGACAT TATCGCGAGC CCATTTATAC CCATATAAAT CAGCATCCAT GTTGGAATTT AATCGCGGCC TAGAGCAAGA CGTTTCCCGT TGAATATGGC TCATAACACC CCTTGTATTA CTGTTTATGT AAGCAGACAG TTTTATTGTT CATGACCAAA ATCCCTTAAC GTGAGTTTTC GTTCCACTGA GCGTCAGACC CCGTAGAAAA GATCAAAGGA TCTTCTTGAG ATCCTTTTTT TCTGCGCGTA ATCTGCTGCT TGCAAACAAA AAAACCACCG CTACCAGCGG TGGTTTGTTT GCCGGATCAA GAGCTACCAA CTCTTTTTCC GAAGGTAACT GGCTTCAGCA GAGCGCAGAT ACCAAATACT GTCCTTCTAG TGTAGCCGTA GTTAGGCCAC CACTTCAAGA ACTCTGTAGC ACCGCCTACA TACCTCGCTC TGCTAATCCT GTTACCAGTG GCTGCTGCCA GTGGCGATAA GTCGTGTCTT ACCGGGTTGG ACTCAAGACG ATAGTTACCG GATAAGGCGC AGCGGTCGGG CTGAACGGGG GGTTCGTGCA CACAGCCCAG CTTGGAGCGA ACGACCTACA CCGAACTGAG ATACCTACAG CGTGAGCTAT GAGAAAGCGC CACGCTTCCC GAAGGGAGAA AGGCGGACAG GTATCCGGTA AGCGGCAGGG TCGGAACAGG AGAGCGCACG AGGGAGCTTC CAGGGGGAAA CGCCTGGTAT CTTTATAGTC CTGTCGGGTT TCGCCACCTC TGACTTGAGC GTCGATTTTT GTGATGCTCG TCAGGGGGGC GGAGCCTATG GAAAAACGCC AGCAACGCGG CCTTTTTACG GTTCCTGGCC TTTTGCTGGC CTTTTGCTCA CATGTTCTTT CCTGCGTTAT CCCCTGATTC TGTGGATAAC CGTATTACCG CCTTTGAGTG AGCTGATACC GCTCGCCGCA GCCGAACGAC CGAGCGCAGC GAGTCAGTGA GCGAGGAAGC GGAAGAGCGC CTGATGCGGT ATTTTCTCCT TACGCATCTG TGCGGTATTT CACACCGCAA TGGTGCACTC TCAGTACAAT CTGCTCTGAT GCCGCATAGT TAAGCCAGTA TACACTCCGC TATCGCTACG TGACTGGGTC ATGGCTGCGC CCCGACACCC GCCAACACCC GCTGACGCGC CCTGACGGGC TTGTCTGCTC CCGGCATCCG CTTACAGACA AGCTGTGACC GTCTCCGGGA GCTGCATGTG TCAGAGGTTT TCACCGTCAT CACCGAAACG CGCGAGGCAG CTGCGGTAAA GCTCATCAGC GTGGTCGTGA AGCGATTCAC AGATGTCTGC CTGTTCATCC GCGTCCAGCT CGTTGAGTTT CTCCAGAAGC GTTAATGTCT GGCTTCTGAT AAAGCGGGCC ATGTTAAGGG CGGTTTTTTC CTGTTTGGTC ACTGATGCCT CCGTGTAAGG GGGATTTCTG TTCATGGGGG TAATGATACC GATGAAACGA GAGAGGATGC TCACGATACG GGTTACTGAT GATGAACATG CCCGGTTACT GGAACGTTGT GAGGGTAAAC AACTGGCGGT ATGGATGCGG CGGGACCAGA GAAAAATCAC TCAGGGTCAA TGCCAGCGCT TCGTTAATAC AGATGTAGGT GTTCCACAGG GTAGCCAGCA GCATCCTGCG ATGCAGATCC GGAACATAAT GGTGCAGGGC GCTGACTTCC GCGTTTCCAG ACTTTACGAA ACACGGAAAC CGAAGACCAT TCATGTTGTT GCTCAGGTCG CAGACGTTTT GCAGCAGCAG TCGCTTCACG TTCGCTCGCG TATCGGTGAT TCATTCTGCT AACCAGTAAG GCAACCCCGC CAGCCTAGCC GGGTCCTCAA CGACAGGAGC ACGATCATGC GCACCCGTGG GGCCGCCATG CCGGCGATAA TGGCCTGCTT CTCGCCGAAA CGTTTGGTGG CGGGACCAGT GACGAAGGCT TGAGCGAGGG CGTGCAAGAT TCCGAATACC GCAAGCGACA GGCCGATCAT CGTCGCGCTC CAGCGAAAGC GGTCCTCGCC GAAAATGACC CAGAGCGCTG CCGGCACCTG TCCTACGAGT TGCATGATAA AGAAGACAGT CATAAGTGCG GCGACGATAG TCATGCCCCG CGCCCACCGG AAGGAGCTGA CTGGGTTGAA GGCTCTCAAG GGCATCGGTC GAGATCCCGG TGCCTAATGA GTGAGCTAAC TTACATTAAT TGCGTTGCGC TCACTGCCCG CTTTCCAGTC GGGAAACCTG TCGTGCCAGC TGCATTAATG AATCGGCCAA CGCGCGGGGA GAGGCGGTTT GCGTATTGGG CGCCAGGGTG GTTTTTCTTT TCACCAGTGA GACGGGCAAC AGCTGATTGC CCTTCACCGC CTGGCCCTGA GAGAGTTGCA GCAAGCGGTC CACGCTGGTT TGCCCCAGCA GGCGAAAATC CTGTTTGATG GTGGTTAACG GCGGGATATA ACATGAGCTG TCTTCGGTAT CGTCGTATCC CACTACCGAG ATATCCGCAC CAACGCGCAG CCCGGACTCG GTAATGGCGC GCATTGCGCC CAGCGCCATC TGATCGTTGG CAACCAGCAT CGCAGTGGGA ACGATGCCCT CATTCAGCAT TTGCATGGTT TGTTGAAAAC CGGACATGGC ACTCCAGTCG CCTTCCCGTT CCGCTATCGG CTGAATTTGA TTGCGAGTGA GATATTTATG CCAGCCAGCC AGACGCAGAC GCGCCGAGAC AGAACTTAAT GGGCCCGCTA ACAGCGCGAT TTGCTGGTGA CCCAATGCGA CCAGATGCTC CACGCCCAGT CGCGTACCGT CTTCATGGGA GAAAATAATA CTGTTGATGG GTGTCTGGTC AGAGACATCA AGAAATAACG CCGGAACATT AGTGCAGGCA GCTTCCACAG CAATGGCATC CTGGTCATCC AGCGGATAGT TAATGATCAG CCCACTGACG CGTTGCGCGA GAAGATTGTG CACCGCCGCT TTACAGGCTT CGACGCCGCT TCGTTCTACC ATCGACACCA CCACGCTGGC ACCCAGTTGA TCGGCGCGAG ATTTAATCGC CGCGACAATT TGCGACGGCG CGTGCAGGGC CAGACTGGAG GTGGCAACGC CAATCAGCAA CGACTGTTTG CCCGCCAGTT GTTGTGCCAC GCGGTTGGGA ATGTAATTCA GCTCCGCCAT CGCCGCTTCC ACTTTTTCCC GCGTTTTCGC AGAAACGTGG CTGGCCTGGT TCACCACGCG GGAAACGGTC TGATAAGAGA CACCGGCATA CTCTGCGACA TCGTATAACG TTACTGGTTT CACATTCACC ACCCTGAATT GACTCTCTTC CGGGCGCTAT CATGCCATAC CGCGAAAGGT TTTGCGCCAT TCGATGGTGT CCGGGATCTC GACGCTCTCC CTTATGCGAC TCCTGCATTA GGAAGCAGCC CAGTAGTAGG TTGAGGCCGT TGAGCACCGC CGCCGCAAGG AATGGTGCAT GCAAGGAGAT GGCGCCCAAC AGTCCCCCGG CCACGGGGCC TGCCACCATA CCCACGCCGA AACAAGCGCT CATGAGCCCG AAGTGGCGAG CCCGATCTTC CCCATCGGTG ATGTCGGCGA TATAGGCGCC AGCAACCGCA CCTGTGGCGC CGGTGATGCC GGCCACGATG CGTCCGGCGT AGAGGATCGA GATCTCGATC CCGCGAAATT AATACGACTC ACTATAGGGG AATTGTGAGC GGATAACAAT TCCCCTCTAG AAATAATTTT GTTTAACTTT AAGAAGGAGA TATACCATGG CTCATCACCA TCACCATCAT ATGGGAACTC TTGAGGCCCA AACACAAGGG CCCGGCAGCA TGCCGGCACT GGTAATTAAA ACGAATGCCA AGTTTACGGA AGAGGAGAAA TCTAAAGCAA CAGAAGAGCT GGGTAATATC GTATCGAAAG TGTTGGGGAA ACCTATTAGT TACGTTATGG TTACCCTTGA AGACGGAGTG GCAGTGCGAT TTGGTGGTTC AGATGAAAAG GCAGCATTTA TGTCTCTGAT GAGCATCGGC GGTCTGAATC GAGCCGTCAA TAAACGCGCT AGCGCCGCGT TGACGAAATG GTTTACCGAT CACGGCTTTC AAGGGGACCG GATATATATT GTGTTTAACC CGAAATCCGC GGAAGATTGG GGTTTTAACG GCGATACCTT CGCATAACTC GAGCACCACC ACCACCACCA CTGAGATCCG GCTGCTAACA AAGCCCGAAA GGAAGCTGAG TTGGCTGCTG CCACCGCTGA GCAATAACTA GCATAACCCC TTGGGGCCTC TAAACGGGTC TTGAGGGGTT TTTTGCTGAA AGGAGGAACT ATATCCGGAT