HosaA.20194.a
IgG H chain
| CENTER ID: | HosaA.20194.a |
| ORGANISM: | Homo sapiens |
| ASSOCIATED DISEASE: | |
| CURRENT STATUS: | in PDB |
| COMMUNITY REQUEST: | True |
| NIH RISK GROUP: | unclassified |
| SELECT AGENT: | False |
| NIH PRIORITY pathogens category: |
unclassified |
Ordering Clones & Proteins
If there are materials available for this target, they will be listed below.
Materials can be ordered from SSGCID using the button in the "order material" column.
Clicking the button will add the material to a virtual cart.
You may order multiple materials at a time at no cost to you, as this contract is funded
by NIAID. When you are ready to place your order, click the "Place Order" link which will
appear in the top right corner of the page after you place your first item in your cart.
Structures
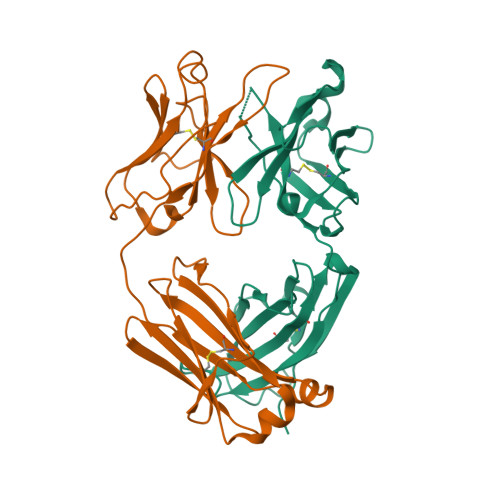
6NB5
DEPOSITED: 12/6/2018
DETERMINATION: XRay
CLONE: HosaA.20194.a.K11.GE42878
PROTEIN: HosaA.20194.a.K11.PE00005
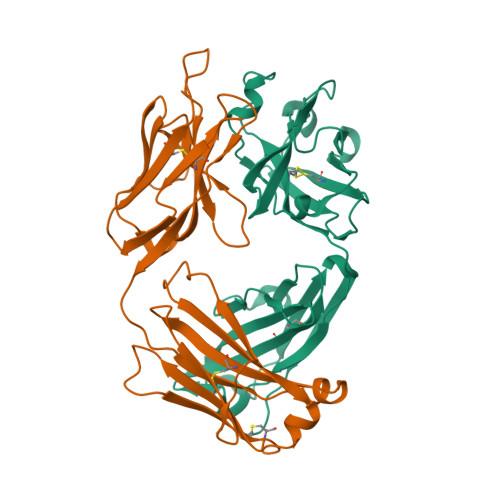
6NB8
DEPOSITED: 12/6/2018
DETERMINATION: XRay
CLONE: HosaA.20194.a.K12.GE42879
PROTEIN: HosaA.20194.a.K12.PE00006
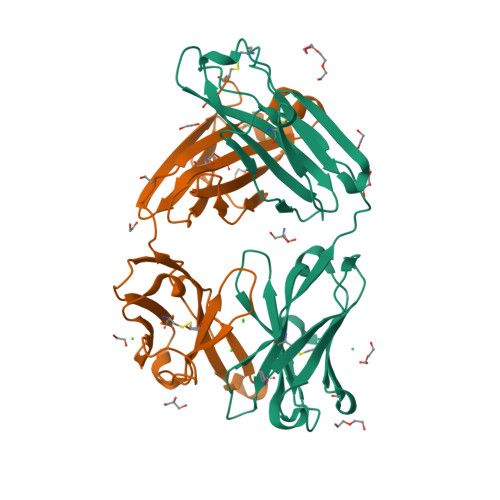
6BLA
DEPOSITED: 11/9/2017
DETERMINATION: XRay
CLONE: HosaA.20194.a.K2.GE43019
PROTEIN: HosaA.20194.a.K2.PE00008
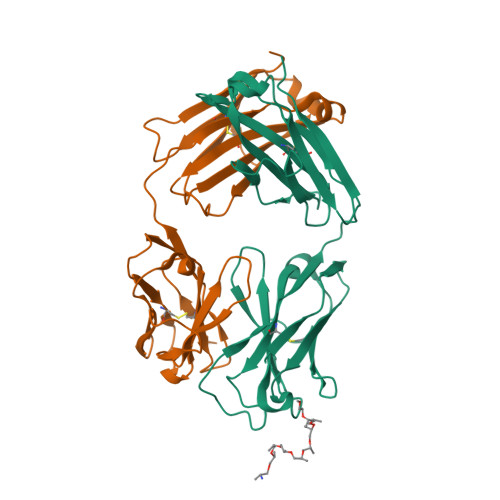
6WS6
DEPOSITED: 4/30/2020
DETERMINATION: XRay
CLONE: HosaA.20194.a.K13.GE43855
PROTEIN: HosaA.20194.a.K13.PE00014
7JXC
DEPOSITED: 8/27/2020
DETERMINATION: XRay
CLONE: HosaA.20194.a.K17.GE44147
PROTEIN: HosaA.20194.a.K17.PE00024
7JXD
DEPOSITED: 8/27/2020
DETERMINATION: XRay
CLONE: HosaA.20194.a.K18.GE44148
PROTEIN: HosaA.20194.a.K18.PE00025
Publications by SSGCID
An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein-Barr virus
Snijder J, McGuire AT, Veesler D, Ortego MS, Weidle C, Stuart AB, Gray MA, McElrath MJ, Pancera M
Immunity - 2018
volume 48, issue 4, pages 799-811.e9
PMID: 29669253; PMCID: PMC5909843
Unexpected receptor functional mimicry elucidates activation of coronavirus fusion
Tortorici MA, Snijder J, Walls AC, Rey FA, Veesler D, Xiong X, Park YJ, Quisp J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Corti D
Cell - 2019
volume 176, issue 5, pages 1026-1039
PMID: 30712865; PMCID: PMC6751136
Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody
Tortorici MA, Walls AC, Park YJ, Cameroni E, Lanzavecchia A, Corti D, Pinto D, Beltramello M, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Case JB, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Diamond M, Fink K
Nature - 2020
volume 583, issue 7815, pages 290-295
PMID: 32422645
Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology
Wall A, Veesler D, Park YJ, Cameroni E, Lanzavecchia A, Corti D, Sallusto F, Tortorici MA, Pinto D, Beltramello M, Jaconi S, Zatta F, De Marco A, Peter A, Guarino B, Havenar-Daughton C, Snell G, Virgin HW, Fink K, Bowen J, Piccoli L, Czudnochowski N, Silacci-Fregni C, Rosen L, Acton O, Minola A, Sprugasci N, Bassi J, Nix J, Mele F, Jovic S, Rodriguez B, Gupta S, Jin F, Piumatti G, Presti G, Pellanda A, Biggiogero M, Tarkowski M, Pizzuto M, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A
Cell - 2020
volume 183, issue 4, pages 1024-1042.e21
PMID: 32991844; PMCID: PMC7494283
External Resources
| RESOURCE | REFERENCE ID |
|---|---|
| UniProt: | S6C4R2 |
Sequences
These sequences are the native gene sequence; sequences of constructs derived from these sequences may differ due to codon optimization or other protocols.
To find the specific sequence of any material you may have ordered, click on the "info" button next to the name of that material.
AA Sequence
MEFGLSWVFL VAILKGVQCE VQLLESGGGL VQPGGSLRLS CAASGFTFSS NAMSWVRQAP GKGLEWVSGV SNSGGDTYYA DSVKGRFAIS RDNSKNTLYL QMNSLRAEDT AVYYCANTLW TVGSKGGFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDKTHTC PPCPAPELLG GTVSLPLPPK TQGHP
NT Sequence
ATGGAGTTTG GGCTGAGCTG GGTTTTTCTT GTGGCTATTT TAAAAGGTGT CCAGTGTGAG GTGCAGCTGT TGGAGTCTGG GGGAGGCTTG GTACAGCCCG GGGGGTCCCT GAGACTCTCC TGTGCAGCCT CTGGATTCAC CTTTAGCAGT AATGCCATGA GCTGGGTCCG CCAGGCTCCA GGGAAGGGGC TGGAGTGGGT CTCAGGTGTT AGTAATAGTG GTGGTGACAC ATACTACGCA GACTCCGTGA AGGGCCGGTT CGCCATCTCC AGAGACAATT CCAAGAACAC GCTCTATCTG CAAATGAACA GCCTGAGAGC CGAGGACACG GCCGTGTATT ACTGTGCGAA TACCCTGTGG ACAGTGGGTA GTAAGGGGGG CTTTGACTAC TGGGGCCAGG GAACCCTGGT CACCGTCTCC TCAGCCTCCA CCAAGGGCCC ATCGGTCTTC CCCCTGGCAC CCTCCTCCAA GAGCACCTCT GGGGGCACAG CGGCCCTGGG CTGCCTGGTC AAGGACTACT TCCCCGAACC GGTGACGGTG TCGTGGAACT CAGGCGCCCT GACCAGCGGC GTGCACACCT TCCCGGCTGT CCTACAGTCC TCAGGACTCT ACTCCCTCAG CAGCGTGGTG ACCGTGCCCT CCAGCAGCTT GGGCACCCAG ACCTACATCT GCAACGTGAA TCACAAGCCC AGCAACACCA AGGTGGACAA GAGAGTTGAG CCCAAATCTT GTGACAAAAC TCACACATGC CCACCGTGCC CAGCACCTGA ACTCCTGGGG GGGACCGTCA GTCTTCCTCT TCCCCCCAAA ACCCAAGGAC ACCCT