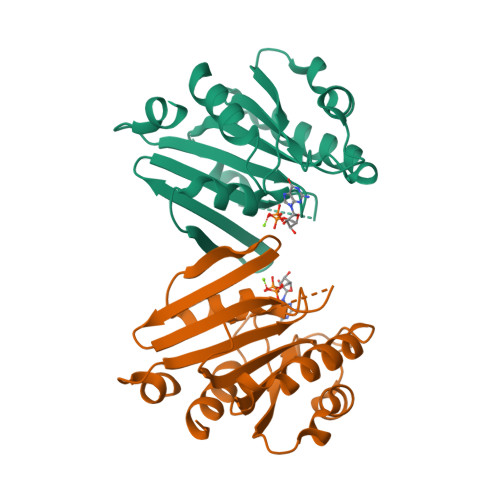
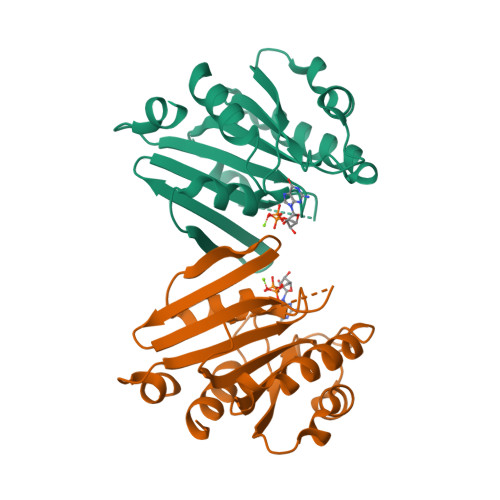
EnhiA.01533.a
ADP-ribosylation factor, putative
| CENTER ID: | EnhiA.01533.a |
| ORGANISM: | Entamoeba histolytica HM-1:IMSS |
| ASSOCIATED DISEASE: | Amoebic dysentery |
| CURRENT STATUS: | in PDB |
| COMMUNITY REQUEST: | False |
| NIH RISK GROUP: | 2 |
| SELECT AGENT: | False |
| NIH PRIORITY pathogens category: |
IIIB |
Ordering Clones & Proteins
If there are materials available for this target, they will be listed below.
Materials can be ordered from SSGCID using the button in the "order material" column.
Clicking the button will add the material to a virtual cart.
You may order multiple materials at a time at no cost to you, as this contract is funded
by NIAID. When you are ready to place your order, click the "Place Order" link which will
appear in the top right corner of the page after you place your first item in your cart.
Proteins
| CENTER REFERENCE ID |
DOMAIN/REGION DESCRIPTION |
INFO | AA START |
AA STOP |
ORDER MATERIAL |
|---|---|---|---|---|---|
| EnhiA.01533.a.A1.PS00671 | Full length( EnhiA.01533.a ) | 1 | 174 | ||
| EnhiA.01533.a.A1.PS00677 | Full length( EnhiA.01533.a ) | 1 | 174 |
Structures
4Y0V
DEPOSITED: 2/6/2015
DETERMINATION: XRay
CLONE: EnhiA.01533.a.A1.GE28433
PROTEIN: EnhiA.01533.a.A1.PS00671
4YLG
DEPOSITED: 3/5/2015
DETERMINATION: XRay
CLONE: EnhiA.01533.a.A1.GE28433
PROTEIN: EnhiA.01533.a.A1.PS00671
Publications by SSGCID
Structure of an ADP-ribosylation factor, ARF1, from Entamoeba histolytica bound to Mg(2+)-GDP.
Clifton MC, Edwards TE, Myler PJ, Sankaran B, Serbzhinskiy DA, Staker BL
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. - 2015
volume 71, issue Pt 5, pages 594-9
PMID: 25945714; PMCID: PMC4427170
External Resources
| RESOURCE | REFERENCE ID |
|---|---|
| EuPathDB: | AmoebaDB:EHI_073470 |
| RefSeq: | XP_656677.1 |
| UniProt: | C4LXL1 |
Sequences
These sequences are the native gene sequence; sequences of constructs derived from these sequences may differ due to codon optimization or other protocols.
To find the specific sequence of any material you may have ordered, click on the "info" button next to the name of that material.
AA Sequence
MGSWLSKLLG KKEMRILMVG LDAAGKTSIL YKLKLGEIVT TIPTIGFNVE TVEYKNISFT VWDVGGQDKI RPLWRHYYQN TQAIIFVVDS NDRDRIGEAR EELMKMLNED EMRNAILLVF ANKHDLPQAM SISEVTEKLG LQTIKNRKWY CQTSCATNGD GLYEGLDWLA DNLK
NT Sequence
ATGGGAAGTT GGTTAAGTAA ATTACTTGGA AAGAAAGAAA TGAGAATTTT GATGGTAGGA CTTGATGCTG CCGGTAAAAC TTCAATTCTT TATAAGTTAA AACTTGGAGA AATTGTCACT ACTATTCCAA CTATTGGATT TAATGTAGAA ACTGTTGAAT ATAAGAATAT TTCCTTTACT GTTTGGGATG TAGGAGGACA AGATAAAATT AGACCATTAT GGAGACATTA TTATCAAAAT ACTCAAGCAA TTATTTTTGT AGTTGATTCT AATGATAGAG ACCGTATTGG AGAAGCACGT GAAGAATTAA TGAAAATGTT AAATGAAGAT GAGATGAGAA ATGCTATTTT GCTTGTTTTT GCTAATAAAC ATGATTTACC ACAAGCTATG TCTATTTCTG AAGTTACAGA AAAACTTGGA TTACAAACAA TTAAGAATCG TAAATGGTAT TGTCAAACAT CATGTGCAAC TAATGGTGAT GGACTTTATG AGGGTCTTGA TTGGCTTGCT GATAATCTCA AA
Details for EnhiA.01533.a.A1.PS00671
| PURIFICATION DATe: | 7/6/2010 |
| CONCENTRATION: | 13.45mg/ml |
| OBSERVED MW: | data unavailable |
| EXPRESSION LEVEL: | High Expression |
| PROTEIN PURIFICATION BUFFER: | 20 mM HEPES, pH 7.0, 300 mM NaCl, 5% glycerol and 1 mM TCEP |
| EXPRESSION HOST: | data unavailable |
| VIAL COUNT (approx.): | 6 |
| VIAL VOLUME: | 200µl |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 100 |
Protocol Notes
notes unavailable
Validated AA Sequence
MAHHHHHHMG TLEAQTQGPG SMGSWLSKLL GKKEMRILMV GLDAAGKTSI LYKLKLGEIV TTIPTIGFNV ETVEYKNISF TVWDVGGQDK IRPLWRHYYQ NTQAIIFVVD SNDRDRIGEA REELMKMLNE DEMRNAILLV FANKHDLPQA MSISEVTEKL GLQTIKNRKW YCQTSCATNG DGLYEGLDWL ADNLK
Validated NT Sequence
annnnnnttt tnnttanctt taagaaggag nnataccatg gctcatcacc atcaccatca tatgggtacc ctggaagctc agacccaggg tcctggttcg atgggaagtt ggttaagtaa attacttgga aagaaagaaa tgagaatttt gatggtagga cttgatgctg ccggtaaaac ttcaattctt tataagttaa aacttggaga aattgtcact actattccaa ctattggatt taatgtagaa actgttgaat ataagaatat ttcctttact gtttgggatg taggaggaca agataaaatt agaccattat ggagacatta ttatcaaaat actcaagcaa ttatttttgt agttgattct aatgatagag accgtattgg agaagcacgt gaagaattaa tgaaaatgtt aaatgaagat gagatgagaa atgctatttt gcttgttttt gctaataaac atgatttacc acaagctatg tctatttctg aagttacaga aaaacttgga ttacaaacaa ttaagaatcg taaatggtat tgtcaaacat catgtgcaac taatggtgat ggactttatg aaggtcttga ttggcttgct gataatctca aataataaac agcacgaaca agttctgcag ccaagcttct cgaggatccg gctgctaaca aagcccgaaa ggaagctgag ttggctgctg ccnnnnnnnn nnntnnnngn ngncgannnn nnnnnncgnn nagnnnnnnc annnnnnnnc natacnnncg annnnnnnnn nnnnnnnngn naannnnnnn nngactctan nannaggnnn nnnnnnncna nnnnnncagn nantncttgn nctananncn acnagnnnnt gatnnacnnc gnnnnnangn nntntncnnn gngatganac annananncn gnntgntgnn ggnctnnnnn nannnngana gnncgngant tnnnnnnnat cnggntcnag nnngnnnnnn gnnngnngnn ncnccnnagn nnnnannggt actnctntnn annnnnngan naanntnnnn anncnnnnng gatcnnnntg ctnnnnnann nnnanggann nnang
Expressed Protein Sequence
MAHHHHHHMG TLEAQTQGPG SMGSWLSKLL GKKEMRILMV GLDAAGKTSI LYKLKLGEIV TTIPTIGFNV ETVEYKNISF TVWDVGGQDK IRPLWRHYYQ NTQAIIFVVD SNDRDRIGEA REELMKMLNE DEMRNAILLV FANKHDLPQA MSISEVTEKL GLQTIKNRKW YCQTSCATNG DGLYEGLDWL ADNLK
Full NT Sequence (Expression Vector + Insert)
TTCTTGAAGA CGAAAGGGCC TCGTGATACG CCTATTTTTA TAGGTTAATG TCATGATAAT AATGGTTTCT TAGACGTCAG GTGGCACTTT TCGGGGAAAT GTGCGCGGAA CCCCTATTTG TTTATTTTTC TAAATACATT CAAATATGTA TCCGCTCATG AGACAATAAC CCTGATAAAT GCTTCAATAA TATTGAAAAA GGAAGAGTAT GAGTATTCAA CATTTCCGTG TCGCCCTTAT TCCCTTTTTT GCGGCATTTT GCCTTCCTGT TTTTGCTCAC CCAGAAACGC TGGTGAAAGT AAAAGATGCT GAAGATCAGT TGGGTGCACG AGTGGGTTAC ATCGAACTGG ATCTCAACAG CGGTAAGATC CTTGAGAGTT TTCGCCCCGA AGAACGTTTT CCAATGATGA GCACTTTTAA AGTTCTGCTA TGTGGCGCGG TATTATCCCG TGTTGACGCC GGGCAAGAGC AACTCGGTCG CCGCATACAC TATTCTCAGA ATGACTTGGT TGAGTACTCA CCAGTCACAG AAAAGCATCT TACGGATGGC ATGACAGTAA GAGAATTATG CAGTGCTGCC ATAACCATGA GTGATAACAC TGCGGCCAAC TTACTTCTGA CAACGATCGG AGGACCGAAG GAGCTAACCG CTTTTTTGCA CAACATGGGG GATCATGTAA CTCGCCTTGA TCGTTGGGAA CCGGAGCTGA ATGAAGCCAT ACCAAACGAC GAGCGTGACA CCACGATGCC TGCAGCAATG GCAACAACGT TGCGCAAACT ATTAACTGGC GAACTACTTA CTCTAGCTTC CCGGCAACAA TTAATAGACT GGATGGAGGC GGATAAAGTT GCAGGACCAC TTCTGCGCTC GGCCCTTCCG GCTGGCTGGT TTATTGCTGA TAAATCTGGA GCCGGTGAGC GTGGGTCTCG CGGTATCATT GCAGCACTGG GGCCAGATGG TAAGCCCTCC CGTATCGTAG TTATCTACAC GACGGGGAGT CAGGCAACTA TGGATGAACG AAATAGACAG ATCGCTGAGA TAGGTGCCTC ACTGATTAAG CATTGGTAAC TGTCAGACCA AGTTTACTCA TATATACTTT AGATTGATTT AAAACTTCAT TTTTAATTTA AAAGGATCTA GGTGAAGATC CTTTTTGATA ATCTCATGAC CAAAATCCCT TAACGTGAGT TTTCGTTCCA CTGAGCGTCA GACCCCGTAG AAAAGATCAA AGGATCTTCT TGAGATCCTT TTTTTCTGCG CGTAATCTGC TGCTTGCAAA CAAAAAAACC ACCGCTACCA GCGGTGGTTT GTTTGCCGGA TCAAGAGCTA CCAACTCTTT TTCCGAAGGT AACTGGCTTC AGCAGAGCGC AGATACCAAA TACTGTCCTT CTAGTGTAGC CGTAGTTAGG CCACCACTTC AAGAACTCTG TAGCACCGCC TACATACCTC GCTCTGCTAA TCCTGTTACC AGTGGCTGCT GCCAGTGGCG ATAAGTCGTG TCTTACCGGG TTGGACTCAA GACGATAGTT ACCGGATAAG GCGCAGCGGT CGGGCTGAAC GGGGGGTTCG TGCACACAGC CCAGCTTGGA GCGAACGACC TACACCGAAC TGAGATACCT ACAGCGTGAG CTATGAGAAA GCGCCACGCT TCCCGAAGGG AGAAAGGCGG ACAGGTATCC GGTAAGCGGC AGGGTCGGAA CAGGAGAGCG CACGAGGGAG CTTCCAGGGG GAAACGCCTG GTATCTTTAT AGTCCTGTCG GGTTTCGCCA CCTCTGACTT GAGCGTCGAT TTTTGTGATG CTCGTCAGGG GGGCGGAGCC TATGGAAAAA CGCCAGCAAC GCGGCCTTTT TACGGTTCCT GGCCTTTTGC TGGCCTTTTG CTCACATGTT CTTTCCTGCG TTATCCCCTG ATTCTGTGGA TAACCGTATT ACCGCCTTTG AGTGAGCTGA TACCGCTCGC CGCAGCCGAA CGACCGAGCG CAGCGAGTCA GTGAGCGAGG AAGCGGAAGA GCGCCTGATG CGGTATTTTC TCCTTACGCA TCTGTGCGGT ATTTCACACC GCATATATGG TGCACTCTCA GTACAATCTG CTCTGATGCC GCATAGTTAA GCCAGTATAC ACTCCGCTAT CGCTACGTGA CTGGGTCATG GCTGCGCCCC GACACCCGCC AACACCCGCT GACGCGCCCT GACGGGCTTG TCTGCTCCCG GCATCCGCTT ACAGACAAGC TGTGACCGTC TCCGGGAGCT GCATGTGTCA GAGGTTTTCA CCGTCATCAC CGAAACGCGC GAGGCAGCTG CGGTAAAGCT CATCAGCGTG GTCGTGAAGC GATTCACAGA TGTCTGCCTG TTCATCCGCG TCCAGCTCGT TGAGTTTCTC CAGAAGCGTT AATGTCTGGC TTCTGATAAA GCGGGCCATG TTAAGGGCGG TTTTTTCCTG TTTGGTCACT GATGCCTCCG TGTAAGGGGG ATTTCTGTTC ATGGGGGTAA TGATACCGAT GAAACGAGAG AGGATGCTCA CGATACGGGT TACTGATGAT GAACATGCCC GGTTACTGGA ACGTTGTGAG GGTAAACAAC TGGCGGTATG GATGCGGCGG GACCAGAGAA AAATCACTCA GGGTCAATGC CAGCGCTTCG TTAATACAGA TGTAGGTGTT CCACAGGGTA GCCAGCAGCA TCCTGCGATG CAGATCCGGA ACATAATGGT GCAGGGCGCT GACTTCCGCG TTTCCAGACT TTACGAAACA CGGAAACCGA AGACCATTCA TGTTGTTGCT CAGGTCGCAG ACGTTTTGCA GCAGCAGTCG CTTCACGTTC GCTCGCGTAT CGGTGATTCA TTCTGCTAAC CAGTAAGGCA ACCCCGCCAG CCTAGCCGGG TCCTCAACGA CAGGAGCACG ATCATGCGCA CCCGTGGCCA GGACCCAACG CTGCCCGAGA TGCGCCGCGT GCGGCTGCTG GAGATGGCGG ACGCGATGGA TATGTTCTGC CAAGGGTTGG TTTGCGCATT CACAGTTCTC CGCAAGAATT GATTGGCTCC AATTCTTGGA GTGGTGAATC CGTTAGCGAG GTGCCGCCGG CTTCCATTCA GGTCGAGGTG GCCCGGCTCC ATGCACCGCG ACGCAACGCG GGGAGGCAGA CAAGGTATAG GGCGGCGCCT ACAATCCATG CCAACCCGTT CCATGTGCTC GCCGAGGCGG CATAAATCGC CGTGACGATC AGCGGTCCAG TGATCGAAGT TAGGCTGGTA AGAGCCGCGA GCGATCCTTG AAGCTGTCCC TGATGGTCGT CATCTACCTG CCTGGACAGC ATGGCCTGCA ACGCGGGCAT CCCGATGCCG CCGGAAGCGA GAAGAATCAT AATGGGGAAG GCCATCCAGC CTCGCGTCGT GAACGCCAGC AAGACGTAGC CCAGCGCGTC GGCCGTAACA ACACCATTTA AATGGAGTGG TTACAAATGG AGTGGTTAAT TAACAACACC ATTTGTCGAC GCTCTCCCTT ATGCGACTCC TGCATTAGGA AGCAGCCCAG TAGTAGGTTG AGGCCGTTGA GCACCGCCGC CGCAAGGAAT GGTGCATGCA AGGAGATGGC GCCCAACAGT CCCCCGGCCA CGGGGCCTGC CACCATACCC ACGCCGAAAC AAGCGCTCAT GAGCCCGAAG TGGCGAGCCC GATCTTCCCC ATCGGTGATG TCGGCGATAT AGGCGCCAGC AACCGCACCT GTGGCGCCGG TGATGCCGGC CACGATGCGT CCGGCGTAGA GGATCGAGAT CTCGATCCCG CGAAATTAAT ACGACTCACT ATAGGGAGAC CACAACGGTT TCCCTCTAGA AATAATTTTG TTTAACTTTA AGAAGGAGAT ATACCATGGC TCATCACCAT CACCATCATA TGGGTACCCT GGAAGCTCAG ACCCAGGGTC CTGGTTCGAT GGGAAGTTGG TTAAGTAAAT TACTTGGAAA GAAAGAAATG AGAATTTTGA TGGTAGGACT TGATGCTGCC GGTAAAACTT CAATTCTTTA TAAGTTAAAA CTTGGAGAAA TTGTCACTAC TATTCCAACT ATTGGATTTA ATGTAGAAAC TGTTGAATAT AAGAATATTT CCTTTACTGT TTGGGATGTA GGAGGACAAG ATAAAATTAG ACCATTATGG AGACATTATT ATCAAAATAC TCAAGCAATT ATTTTTGTAG TTGATTCTAA TGATAGAGAC CGTATTGGAG AAGCACGTGA AGAATTAATG AAAATGTTAA ATGAAGATGA GATGAGAAAT GCTATTTTGC TTGTTTTTGC TAATAAACAT GATTTACCAC AAGCTATGTC TATTTCTGAA GTTACAGAAA AACTTGGATT ACAAACAATT AAGAATCGTA AATGGTATTG TCAAACATCA TGTGCAACTA ATGGTGATGG ACTTTATGAG GGTCTTGATT GGCTTGCTGA TAATCTCAAA AAACAGCACG AACAAGTTCT GCAGCCAAGC TTCTCGAGGA TCCGGCTGCT AACAAAGCCC GAAAGGAAGC TGAGTTGGCT GCTGCCACCG CTGAGCAATA ACTAGCATAA CCCCTTGGGG CCTCTAAACG GGTCTTGAGG GGTTTTTTGC TGAAAGGAGG AACTATATCC GGATATCCAC AGGACGGGTG TGGTCGCCAT GATCGCGTAG TCGATAGTGG CTCCAAGTAG CGAAGCGAGC AGGACTGGGC GGCGGCCAAA GCGGTCGGAC AGTGCTCCGA GAACGGGTGC GCATAGAAAT TGCATCAACG CATATAGCGC TAGCAGCACG CCATAGTGAC TGGCGATGCT GTCGGAATGG ACGATATCCC GCAAGAGGCC CGGCAGTACC GGCATAACCA AGCCTATGCC TACAGCATCC AGGGTGACGG TGCCGAGGAT GACGATGAGC GCATTGTTAG ATTTCATACA CGGTGCCTGA CTGCGTTAGC AATTTAACTG TGATAAACTA CCGCATTAAA GCTTATCGAT GATAAGCTGT CAAACATGAG AA
Details for EnhiA.01533.a.A1.PS00677
| PURIFICATION DATe: | 7/6/2010 |
| CONCENTRATION: | 54.37mg/ml |
| OBSERVED MW: | data unavailable |
| EXPRESSION LEVEL: | High Expression |
| PROTEIN PURIFICATION BUFFER: | 20 mM HEPES, pH 7.0, 300 mM NaCl, 5% glycerol and 1 mM TCEP |
| EXPRESSION HOST: | data unavailable |
| VIAL COUNT (approx.): | 5 |
| VIAL VOLUME: | 200µl |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 100 |
Protocol Notes
notes unavailable
Validated AA Sequence
MAHHHHHHMG TLEAQTQGPG SMGSWLSKLL GKKEMRILMV GLDAAGKTSI LYKLKLGEIV TTIPTIGFNV ETVEYKNISF TVWDVGGQDK IRPLWRHYYQ NTQAIIFVVD SNDRDRIGEA REELMKMLNE DEMRNAILLV FANKHDLPQA MSISEVTEKL GLQTIKNRKW YCQTSCATNG DGLYEGLDWL ADNLK
Validated NT Sequence
annnnnnttt tnnttanctt taagaaggag nnataccatg gctcatcacc atcaccatca tatgggtacc ctggaagctc agacccaggg tcctggttcg atgggaagtt ggttaagtaa attacttgga aagaaagaaa tgagaatttt gatggtagga cttgatgctg ccggtaaaac ttcaattctt tataagttaa aacttggaga aattgtcact actattccaa ctattggatt taatgtagaa actgttgaat ataagaatat ttcctttact gtttgggatg taggaggaca agataaaatt agaccattat ggagacatta ttatcaaaat actcaagcaa ttatttttgt agttgattct aatgatagag accgtattgg agaagcacgt gaagaattaa tgaaaatgtt aaatgaagat gagatgagaa atgctatttt gcttgttttt gctaataaac atgatttacc acaagctatg tctatttctg aagttacaga aaaacttgga ttacaaacaa ttaagaatcg taaatggtat tgtcaaacat catgtgcaac taatggtgat ggactttatg aaggtcttga ttggcttgct gataatctca aataataaac agcacgaaca agttctgcag ccaagcttct cgaggatccg gctgctaaca aagcccgaaa ggaagctgag ttggctgctg ccnnnnnnnn nnntnnnngn ngncgannnn nnnnnncgnn nagnnnnnnc annnnnnnnc natacnnncg annnnnnnnn nnnnnnnngn naannnnnnn nngactctan nannaggnnn nnnnnnncna nnnnnncagn nantncttgn nctananncn acnagnnnnt gatnnacnnc gnnnnnangn nntntncnnn gngatganac annananncn gnntgntgnn ggnctnnnnn nannnngana gnncgngant tnnnnnnnat cnggntcnag nnngnnnnnn gnnngnngnn ncnccnnagn nnnnannggt actnctntnn annnnnngan naanntnnnn anncnnnnng gatcnnnntg ctnnnnnann nnnanggann nnang
Expressed Protein Sequence
MAHHHHHHMG TLEAQTQGPG SMGSWLSKLL GKKEMRILMV GLDAAGKTSI LYKLKLGEIV TTIPTIGFNV ETVEYKNISF TVWDVGGQDK IRPLWRHYYQ NTQAIIFVVD SNDRDRIGEA REELMKMLNE DEMRNAILLV FANKHDLPQA MSISEVTEKL GLQTIKNRKW YCQTSCATNG DGLYEGLDWL ADNLK
Full NT Sequence (Expression Vector + Insert)
TTCTTGAAGA CGAAAGGGCC TCGTGATACG CCTATTTTTA TAGGTTAATG TCATGATAAT AATGGTTTCT TAGACGTCAG GTGGCACTTT TCGGGGAAAT GTGCGCGGAA CCCCTATTTG TTTATTTTTC TAAATACATT CAAATATGTA TCCGCTCATG AGACAATAAC CCTGATAAAT GCTTCAATAA TATTGAAAAA GGAAGAGTAT GAGTATTCAA CATTTCCGTG TCGCCCTTAT TCCCTTTTTT GCGGCATTTT GCCTTCCTGT TTTTGCTCAC CCAGAAACGC TGGTGAAAGT AAAAGATGCT GAAGATCAGT TGGGTGCACG AGTGGGTTAC ATCGAACTGG ATCTCAACAG CGGTAAGATC CTTGAGAGTT TTCGCCCCGA AGAACGTTTT CCAATGATGA GCACTTTTAA AGTTCTGCTA TGTGGCGCGG TATTATCCCG TGTTGACGCC GGGCAAGAGC AACTCGGTCG CCGCATACAC TATTCTCAGA ATGACTTGGT TGAGTACTCA CCAGTCACAG AAAAGCATCT TACGGATGGC ATGACAGTAA GAGAATTATG CAGTGCTGCC ATAACCATGA GTGATAACAC TGCGGCCAAC TTACTTCTGA CAACGATCGG AGGACCGAAG GAGCTAACCG CTTTTTTGCA CAACATGGGG GATCATGTAA CTCGCCTTGA TCGTTGGGAA CCGGAGCTGA ATGAAGCCAT ACCAAACGAC GAGCGTGACA CCACGATGCC TGCAGCAATG GCAACAACGT TGCGCAAACT ATTAACTGGC GAACTACTTA CTCTAGCTTC CCGGCAACAA TTAATAGACT GGATGGAGGC GGATAAAGTT GCAGGACCAC TTCTGCGCTC GGCCCTTCCG GCTGGCTGGT TTATTGCTGA TAAATCTGGA GCCGGTGAGC GTGGGTCTCG CGGTATCATT GCAGCACTGG GGCCAGATGG TAAGCCCTCC CGTATCGTAG TTATCTACAC GACGGGGAGT CAGGCAACTA TGGATGAACG AAATAGACAG ATCGCTGAGA TAGGTGCCTC ACTGATTAAG CATTGGTAAC TGTCAGACCA AGTTTACTCA TATATACTTT AGATTGATTT AAAACTTCAT TTTTAATTTA AAAGGATCTA GGTGAAGATC CTTTTTGATA ATCTCATGAC CAAAATCCCT TAACGTGAGT TTTCGTTCCA CTGAGCGTCA GACCCCGTAG AAAAGATCAA AGGATCTTCT TGAGATCCTT TTTTTCTGCG CGTAATCTGC TGCTTGCAAA CAAAAAAACC ACCGCTACCA GCGGTGGTTT GTTTGCCGGA TCAAGAGCTA CCAACTCTTT TTCCGAAGGT AACTGGCTTC AGCAGAGCGC AGATACCAAA TACTGTCCTT CTAGTGTAGC CGTAGTTAGG CCACCACTTC AAGAACTCTG TAGCACCGCC TACATACCTC GCTCTGCTAA TCCTGTTACC AGTGGCTGCT GCCAGTGGCG ATAAGTCGTG TCTTACCGGG TTGGACTCAA GACGATAGTT ACCGGATAAG GCGCAGCGGT CGGGCTGAAC GGGGGGTTCG TGCACACAGC CCAGCTTGGA GCGAACGACC TACACCGAAC TGAGATACCT ACAGCGTGAG CTATGAGAAA GCGCCACGCT TCCCGAAGGG AGAAAGGCGG ACAGGTATCC GGTAAGCGGC AGGGTCGGAA CAGGAGAGCG CACGAGGGAG CTTCCAGGGG GAAACGCCTG GTATCTTTAT AGTCCTGTCG GGTTTCGCCA CCTCTGACTT GAGCGTCGAT TTTTGTGATG CTCGTCAGGG GGGCGGAGCC TATGGAAAAA CGCCAGCAAC GCGGCCTTTT TACGGTTCCT GGCCTTTTGC TGGCCTTTTG CTCACATGTT CTTTCCTGCG TTATCCCCTG ATTCTGTGGA TAACCGTATT ACCGCCTTTG AGTGAGCTGA TACCGCTCGC CGCAGCCGAA CGACCGAGCG CAGCGAGTCA GTGAGCGAGG AAGCGGAAGA GCGCCTGATG CGGTATTTTC TCCTTACGCA TCTGTGCGGT ATTTCACACC GCATATATGG TGCACTCTCA GTACAATCTG CTCTGATGCC GCATAGTTAA GCCAGTATAC ACTCCGCTAT CGCTACGTGA CTGGGTCATG GCTGCGCCCC GACACCCGCC AACACCCGCT GACGCGCCCT GACGGGCTTG TCTGCTCCCG GCATCCGCTT ACAGACAAGC TGTGACCGTC TCCGGGAGCT GCATGTGTCA GAGGTTTTCA CCGTCATCAC CGAAACGCGC GAGGCAGCTG CGGTAAAGCT CATCAGCGTG GTCGTGAAGC GATTCACAGA TGTCTGCCTG TTCATCCGCG TCCAGCTCGT TGAGTTTCTC CAGAAGCGTT AATGTCTGGC TTCTGATAAA GCGGGCCATG TTAAGGGCGG TTTTTTCCTG TTTGGTCACT GATGCCTCCG TGTAAGGGGG ATTTCTGTTC ATGGGGGTAA TGATACCGAT GAAACGAGAG AGGATGCTCA CGATACGGGT TACTGATGAT GAACATGCCC GGTTACTGGA ACGTTGTGAG GGTAAACAAC TGGCGGTATG GATGCGGCGG GACCAGAGAA AAATCACTCA GGGTCAATGC CAGCGCTTCG TTAATACAGA TGTAGGTGTT CCACAGGGTA GCCAGCAGCA TCCTGCGATG CAGATCCGGA ACATAATGGT GCAGGGCGCT GACTTCCGCG TTTCCAGACT TTACGAAACA CGGAAACCGA AGACCATTCA TGTTGTTGCT CAGGTCGCAG ACGTTTTGCA GCAGCAGTCG CTTCACGTTC GCTCGCGTAT CGGTGATTCA TTCTGCTAAC CAGTAAGGCA ACCCCGCCAG CCTAGCCGGG TCCTCAACGA CAGGAGCACG ATCATGCGCA CCCGTGGCCA GGACCCAACG CTGCCCGAGA TGCGCCGCGT GCGGCTGCTG GAGATGGCGG ACGCGATGGA TATGTTCTGC CAAGGGTTGG TTTGCGCATT CACAGTTCTC CGCAAGAATT GATTGGCTCC AATTCTTGGA GTGGTGAATC CGTTAGCGAG GTGCCGCCGG CTTCCATTCA GGTCGAGGTG GCCCGGCTCC ATGCACCGCG ACGCAACGCG GGGAGGCAGA CAAGGTATAG GGCGGCGCCT ACAATCCATG CCAACCCGTT CCATGTGCTC GCCGAGGCGG CATAAATCGC CGTGACGATC AGCGGTCCAG TGATCGAAGT TAGGCTGGTA AGAGCCGCGA GCGATCCTTG AAGCTGTCCC TGATGGTCGT CATCTACCTG CCTGGACAGC ATGGCCTGCA ACGCGGGCAT CCCGATGCCG CCGGAAGCGA GAAGAATCAT AATGGGGAAG GCCATCCAGC CTCGCGTCGT GAACGCCAGC AAGACGTAGC CCAGCGCGTC GGCCGTAACA ACACCATTTA AATGGAGTGG TTACAAATGG AGTGGTTAAT TAACAACACC ATTTGTCGAC GCTCTCCCTT ATGCGACTCC TGCATTAGGA AGCAGCCCAG TAGTAGGTTG AGGCCGTTGA GCACCGCCGC CGCAAGGAAT GGTGCATGCA AGGAGATGGC GCCCAACAGT CCCCCGGCCA CGGGGCCTGC CACCATACCC ACGCCGAAAC AAGCGCTCAT GAGCCCGAAG TGGCGAGCCC GATCTTCCCC ATCGGTGATG TCGGCGATAT AGGCGCCAGC AACCGCACCT GTGGCGCCGG TGATGCCGGC CACGATGCGT CCGGCGTAGA GGATCGAGAT CTCGATCCCG CGAAATTAAT ACGACTCACT ATAGGGAGAC CACAACGGTT TCCCTCTAGA AATAATTTTG TTTAACTTTA AGAAGGAGAT ATACCATGGC TCATCACCAT CACCATCATA TGGGTACCCT GGAAGCTCAG ACCCAGGGTC CTGGTTCGAT GGGAAGTTGG TTAAGTAAAT TACTTGGAAA GAAAGAAATG AGAATTTTGA TGGTAGGACT TGATGCTGCC GGTAAAACTT CAATTCTTTA TAAGTTAAAA CTTGGAGAAA TTGTCACTAC TATTCCAACT ATTGGATTTA ATGTAGAAAC TGTTGAATAT AAGAATATTT CCTTTACTGT TTGGGATGTA GGAGGACAAG ATAAAATTAG ACCATTATGG AGACATTATT ATCAAAATAC TCAAGCAATT ATTTTTGTAG TTGATTCTAA TGATAGAGAC CGTATTGGAG AAGCACGTGA AGAATTAATG AAAATGTTAA ATGAAGATGA GATGAGAAAT GCTATTTTGC TTGTTTTTGC TAATAAACAT GATTTACCAC AAGCTATGTC TATTTCTGAA GTTACAGAAA AACTTGGATT ACAAACAATT AAGAATCGTA AATGGTATTG TCAAACATCA TGTGCAACTA ATGGTGATGG ACTTTATGAG GGTCTTGATT GGCTTGCTGA TAATCTCAAA AAACAGCACG AACAAGTTCT GCAGCCAAGC TTCTCGAGGA TCCGGCTGCT AACAAAGCCC GAAAGGAAGC TGAGTTGGCT GCTGCCACCG CTGAGCAATA ACTAGCATAA CCCCTTGGGG CCTCTAAACG GGTCTTGAGG GGTTTTTTGC TGAAAGGAGG AACTATATCC GGATATCCAC AGGACGGGTG TGGTCGCCAT GATCGCGTAG TCGATAGTGG CTCCAAGTAG CGAAGCGAGC AGGACTGGGC GGCGGCCAAA GCGGTCGGAC AGTGCTCCGA GAACGGGTGC GCATAGAAAT TGCATCAACG CATATAGCGC TAGCAGCACG CCATAGTGAC TGGCGATGCT GTCGGAATGG ACGATATCCC GCAAGAGGCC CGGCAGTACC GGCATAACCA AGCCTATGCC TACAGCATCC AGGGTGACGG TGCCGAGGAT GACGATGAGC GCATTGTTAG ATTTCATACA CGGTGCCTGA CTGCGTTAGC AATTTAACTG TGATAAACTA CCGCATTAAA GCTTATCGAT GATAAGCTGT CAAACATGAG AA