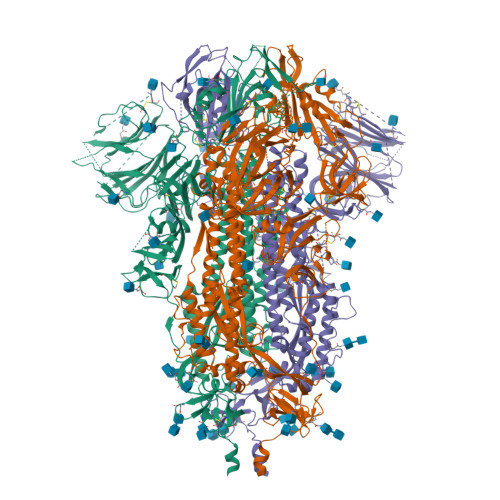
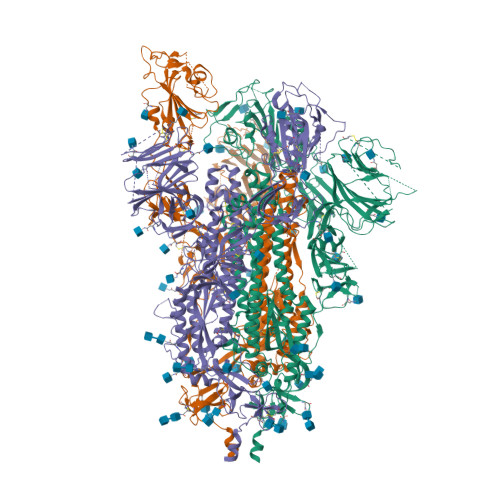
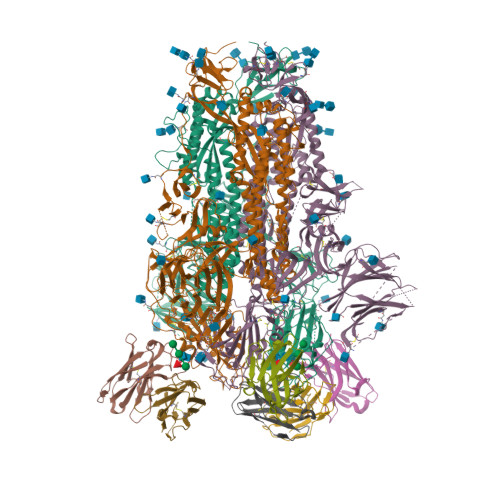
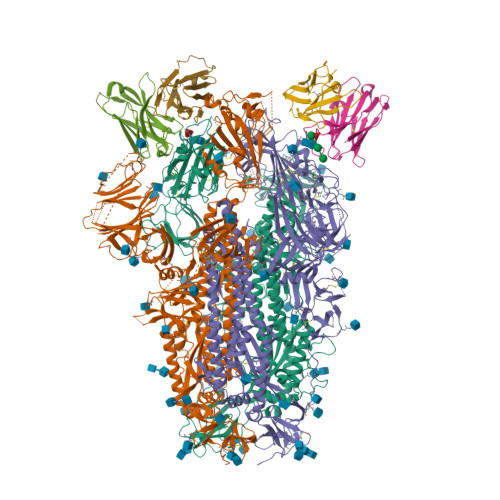
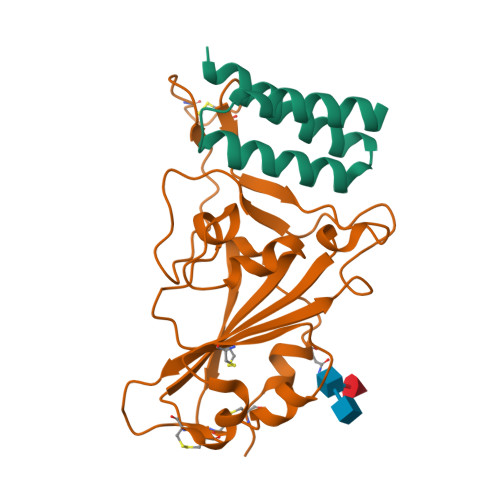
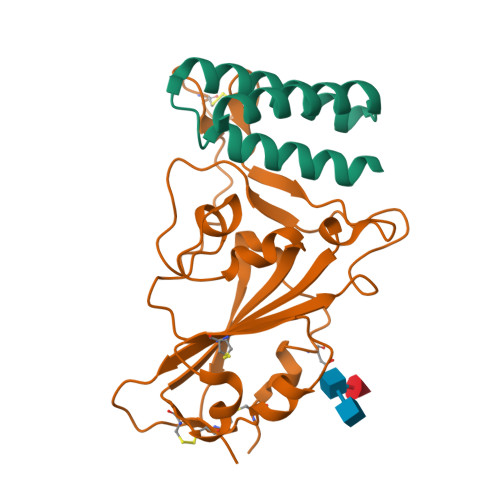
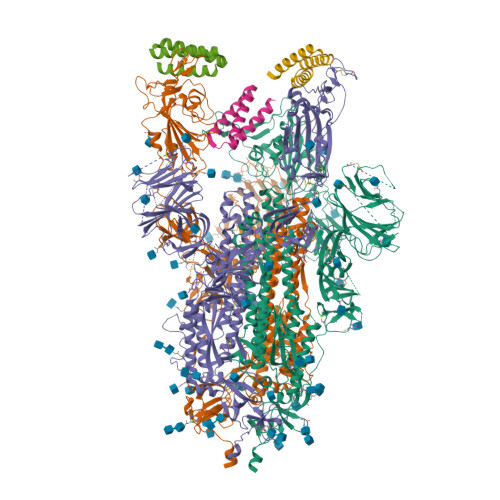
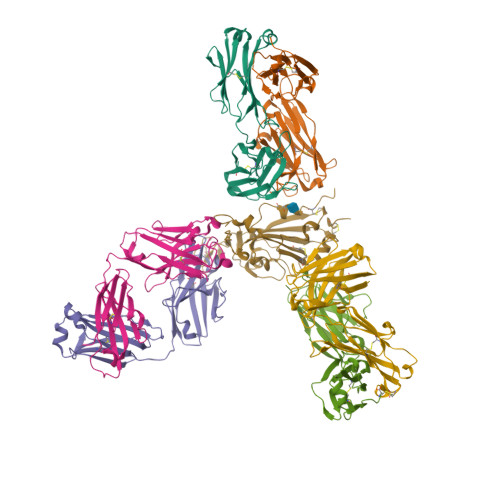
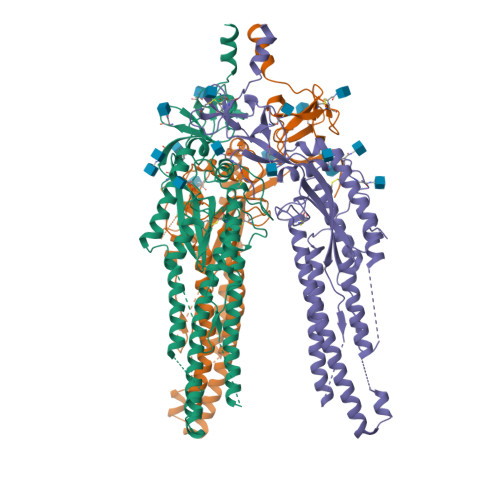
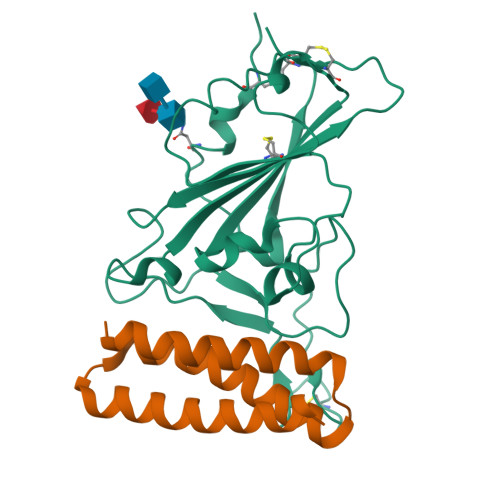
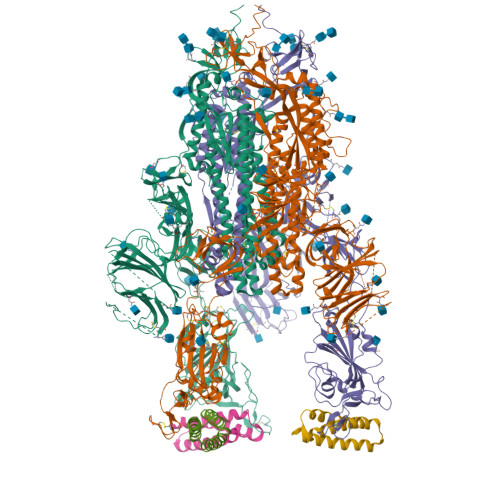
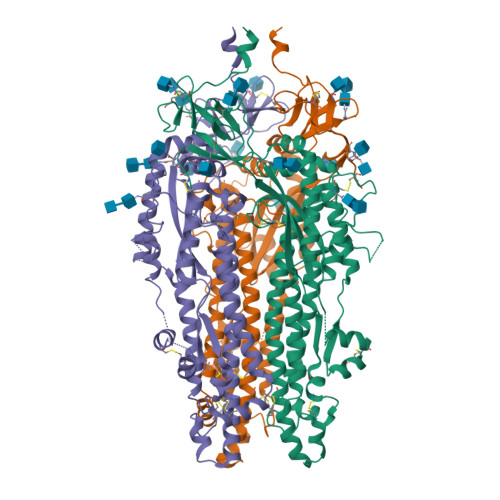
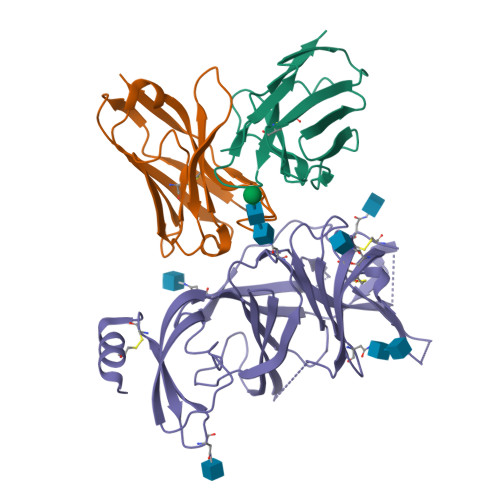
BewuA.18655.a
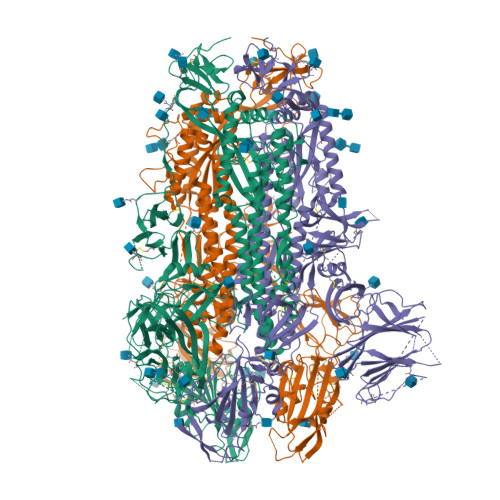
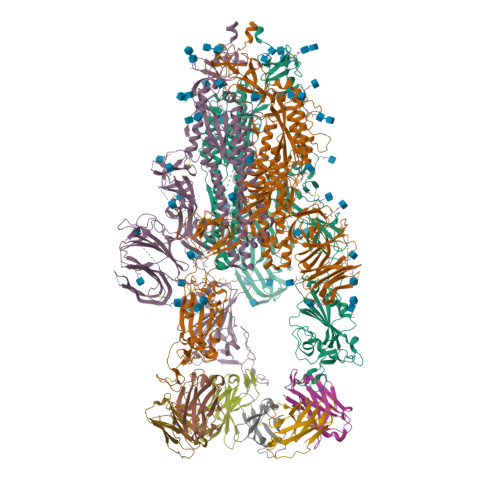
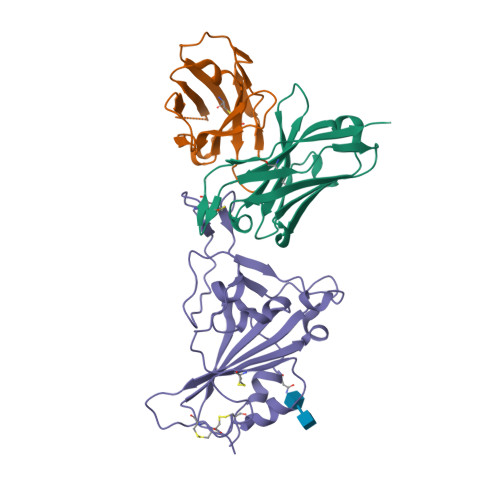
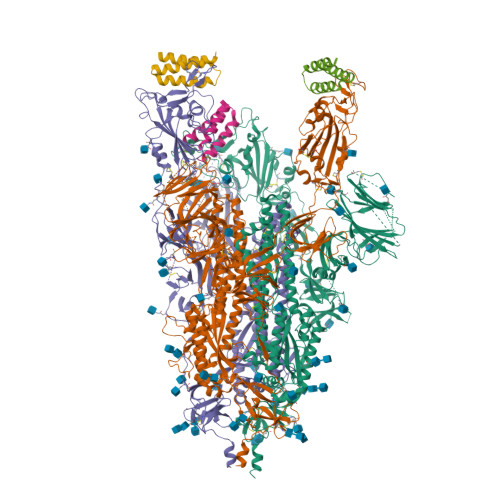
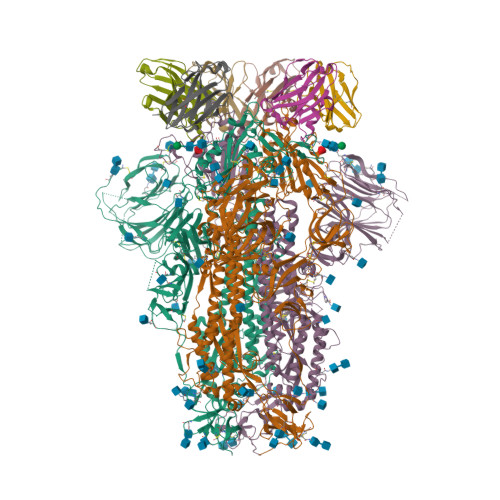
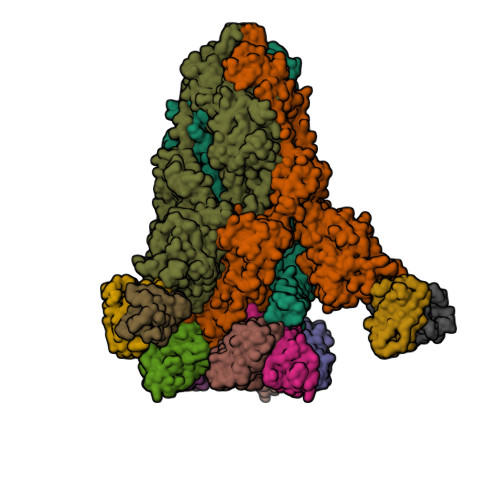
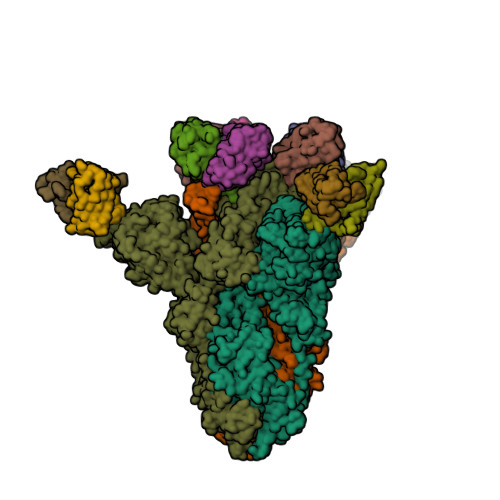
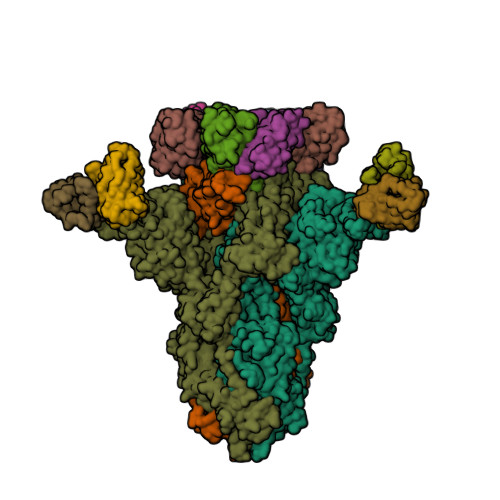
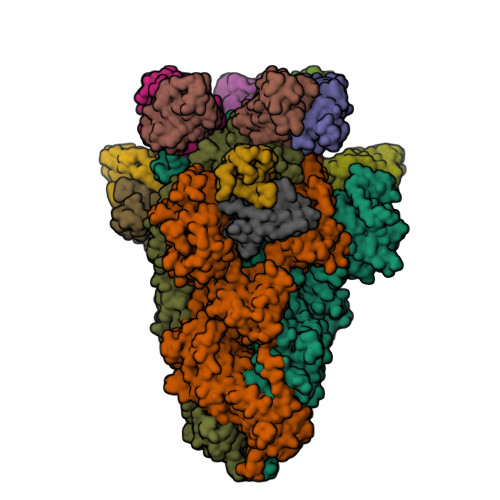
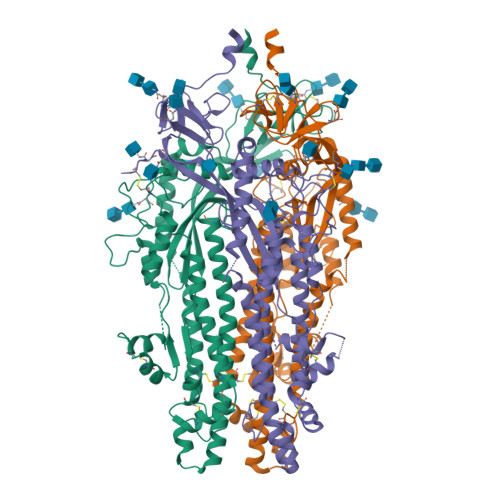
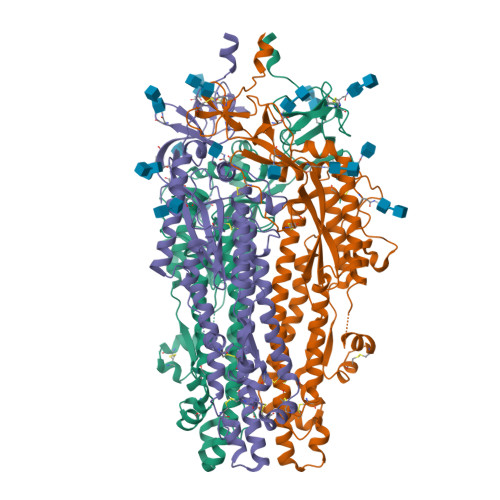
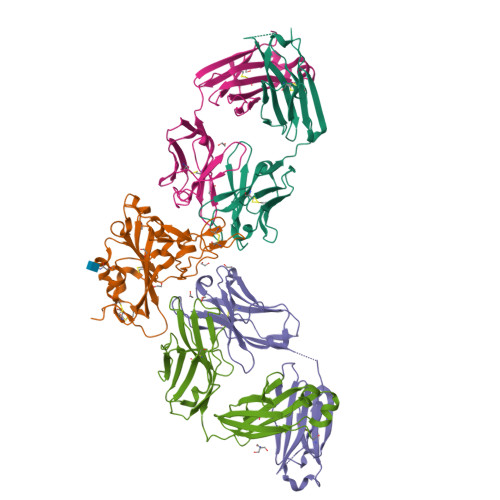
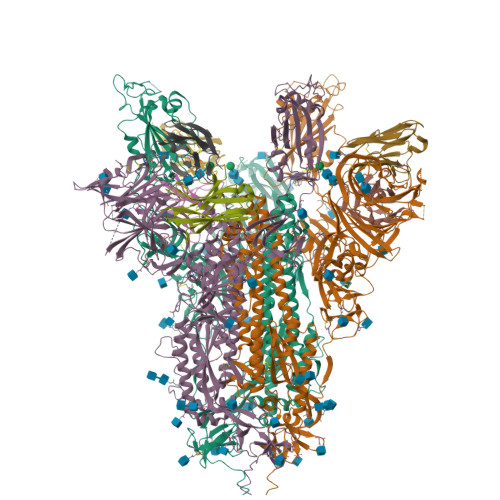
Spike glycoprotein (S)
| CENTER ID: | BewuA.18655.a |
| ORGANISM: | Betacoronavirus SARS-CoV-2 (2019-nCoV) (COVID-19) isolate Wuhan-Hu-1 |
| ASSOCIATED DISEASE: | |
| CURRENT STATUS: | in PDB |
| COMMUNITY REQUEST: | True |
| NIH RISK GROUP: | 2 |
| SELECT AGENT: | False |
| NIH PRIORITY pathogens category: |
IIIC |
Ordering Clones & Proteins
If there are materials available for this target, they will be listed below.
Materials can be ordered from SSGCID using the button in the "order material" column.
Clicking the button will add the material to a virtual cart.
You may order multiple materials at a time at no cost to you, as this contract is funded
by NIAID. When you are ready to place your order, click the "Place Order" link which will
appear in the top right corner of the page after you place your first item in your cart.
Structures
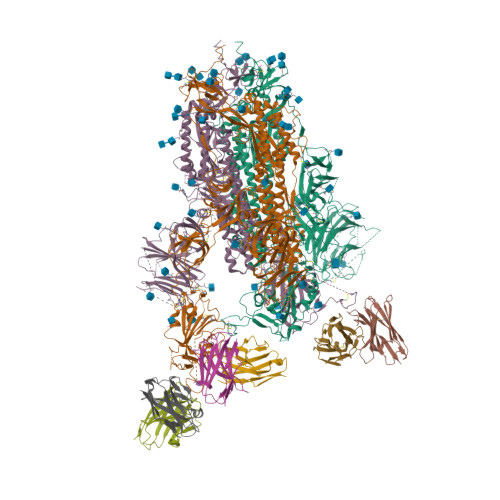
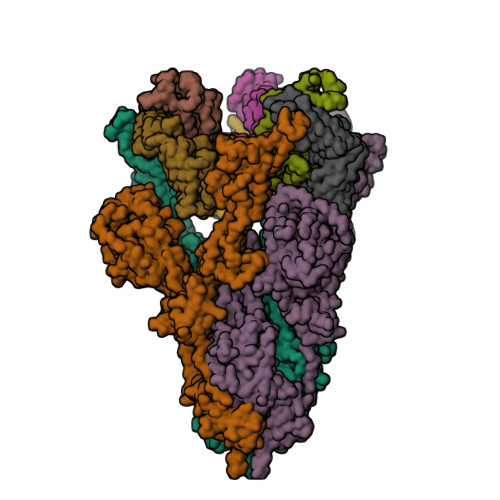
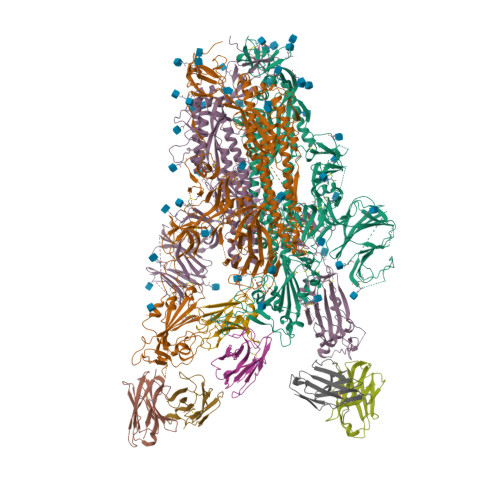
6VXX
DEPOSITED: 2/25/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.MG1.GE43647
PROTEIN: BewuA.18655.a.MG1.PE00013
6VYB
DEPOSITED: 2/25/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.MG1.GE43647
PROTEIN: BewuA.18655.a.MG1.PE00013
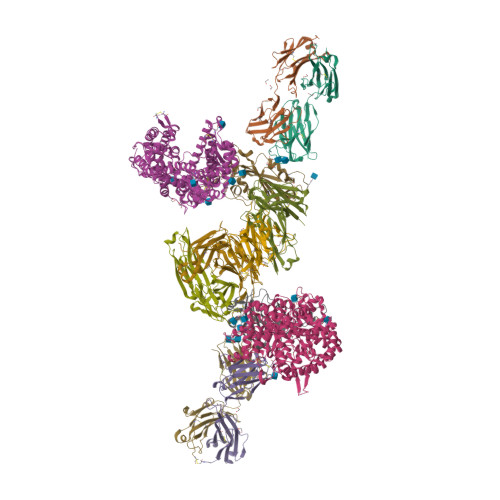
6WPS
DEPOSITED: 4/27/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.K14.GE43854
PROTEIN: BewuA.18655.a.K14.PE00015
6WPT
DEPOSITED: 4/27/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.K14.GE43854
PROTEIN: BewuA.18655.a.K14.PE00015
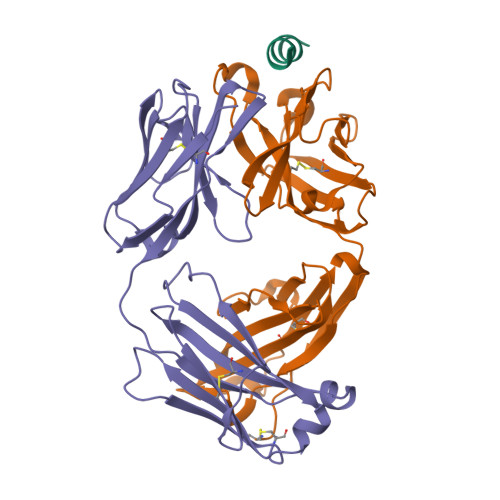
6X79
DEPOSITED: 5/29/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.TZ31.GE43861
PROTEIN: BewuA.18655.a.TZ31.PE00016
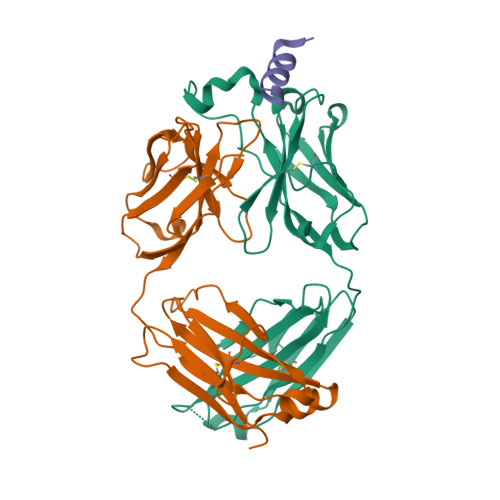
7K4N
DEPOSITED: 9/15/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00017
7K45
DEPOSITED: 9/14/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00017
7JZL
DEPOSITED: 9/2/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00018
7JZU
DEPOSITED: 9/2/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00018
7JZM
DEPOSITED: 9/2/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00019
7JZN
DEPOSITED: 9/2/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00019
7JX3
DEPOSITED: 8/26/2020
DETERMINATION: XRay
CLONE: BewuA.18655.a.K2.GE44140
PROTEIN: BewuA.18655.a.K2.PE00020
7K43
DEPOSITED: 9/14/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PE00021
7JVA
DEPOSITED: 8/20/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PS38661
7JVC
DEPOSITED: 8/20/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PS38661
7JV6
DEPOSITED: 8/20/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PE00022
7JV4
DEPOSITED: 8/20/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PE00022
7JV2
DEPOSITED: 8/20/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PE00022
7JW0
DEPOSITED: 8/24/2020
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PK31.GE44142
PROTEIN: BewuA.18655.a.PK31.PE00023
7LXW
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00033
7LXY
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00034
7LXX
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00035
7LXZ
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00036
7LY0
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00037
7LY2
DEPOSITED: 3/5/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44141
PROTEIN: BewuA.18655.a.PJ41.PE00038
7LY3
DEPOSITED: 3/5/2021
DETERMINATION: XRay
CLONE: BewuA.18655.a.K41.GE44378
PROTEIN: BewuA.18655.a.K41.PE00039
7RA8
DEPOSITED: 6/30/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44489
PROTEIN: BewuA.18655.a.PJ41.PE00044
7RAL
DEPOSITED: 7/1/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44489
PROTEIN: BewuA.18655.a.PJ41.PE00044
7N8H
DEPOSITED: 6/14/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.UP41.GE44484
PROTEIN: BewuA.18655.a.UP41.PE00042
7N8I
DEPOSITED: 6/14/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.UP41.GE44484
PROTEIN: BewuA.18655.a.UP41.PE00042
7R7N
DEPOSITED: 6/25/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44481
PROTEIN: BewuA.18655.a.PJ41.PE00043
7TAS
DEPOSITED: 12/21/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44703
PROTEIN: BewuA.18655.a.PJ41.PE00050
7TAT
DEPOSITED: 12/21/2021
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44703
PROTEIN: BewuA.18655.a.PJ41.PE00050
8DF5
DEPOSITED: 6/21/2022
DETERMINATION: XRay
CLONE: BewuA.18655.a.K2.GE44140
PROTEIN: BewuA.18655.a.K2.PE00060
7SKZ
DEPOSITED: 10/22/2021
DETERMINATION: XRay
CLONE: BewuA.18655.a.K51.GE44809
PROTEIN: BewuA.18655.a.K51.PE00061
7SL5
DEPOSITED: 10/22/2021
DETERMINATION: XRay
CLONE: BewuA.18655.a.K52.GE44810
PROTEIN: BewuA.18655.a.K52.PE00062
8DYA
DEPOSITED: 8/3/2022
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.UP42.GE44911
PROTEIN: BewuA.18655.a.UP42.PS38708
7UHB
DEPOSITED: 3/26/2022
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44703
PROTEIN: BewuA.18655.a.PJ41.PE00050
7UHC
DEPOSITED: 3/26/2022
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.PJ41.GE44703
PROTEIN: BewuA.18655.a.PJ41.PE00050
8VQ9
DEPOSITED: 1/18/2024
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.K61.GE45600
PROTEIN: BewuA.18655.a.K61.PE00080
8VQA
DEPOSITED: 1/18/2024
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.K62.GE45601
PROTEIN: BewuA.18655.a.K62.PE00081
8VQB
DEPOSITED: 1/18/2024
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.K63.GE45602
PROTEIN: BewuA.18655.a.K63.PE00082
8S6M
DEPOSITED: 2/28/2024
DETERMINATION: XRay
CLONE: BewuA.18655.a.ME21.GE45859
PROTEIN: BewuA.18655.a.ME21.PE00091
9C44
DEPOSITED: 6/3/2024
DETERMINATION: CryoEM
CLONE: BewuA.18655.a.UP43.GE45875
PROTEIN: BewuA.18655.a.UP43.PE00096
Molecular models
| METHOD | RESULTS |
|---|---|
| comparative modelling | Robetta_15652 |
Publications by SSGCID
Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein
Wall A, Tortorici MA, Walls AC, McGuire AT, Veesler D, Park YJ
Cell - 2020
volume 181, issue 2, pages 281-292.e6
PMID: 32155444; PMCID: PMC7102599
Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody
Tortorici MA, Walls AC, Park YJ, Cameroni E, Lanzavecchia A, Corti D, Pinto D, Beltramello M, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Case JB, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Diamond M, Fink K
Nature - 2020
volume 583, issue 7815, pages 290-295
PMID: 32422645
Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation
Walls AC, Veesler D, Corti D, McCallum M, Bowen J
Nat. Struct. Mol. Biol. - 2020
volume 27, issue 10, pages 942-949
PMID: 32753755; PMCID: PMC7541350
De novo design of picomolar SARS-CoV-2 miniprotein inhibitors
Stewart L, Walls AC, Park YJ, Carter L, Baker D, Case JB, Chen R, Diamond M, Kozodoy L, Miller L, Coventry B, Goreshnik I, Cao L
Science - 2020
volume 370, issue 6515, pages 426-431
PMID: 32907861; PMCID: PMC7857403
Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2
Chen C, Walls AC, Fiala B, Hodge E, Carter L, Lee KK, King NP, Ravichandran R, Chu H, Schafer A, Wrenn S, Pham M, Murphy M, Tse L, Shehata L, O'Connor M, Navarro M, Miranda M, Kraft J, Ogohara C, Palser A, Chalk S, Lee E, Kepl E, Chow C, Sydeman C, Brown B, Fuller J, Dinnon K, Gralinski L, Leist S, Gully K, Lewis T, Guttman M, Fuller D, Baric R, Kellam P, Pepper M, Sheahan T
Cell - 2020
volume 183, issue 5, pages 1367-1382.e17
PMID: 33160446; PMCID: PMC7604136
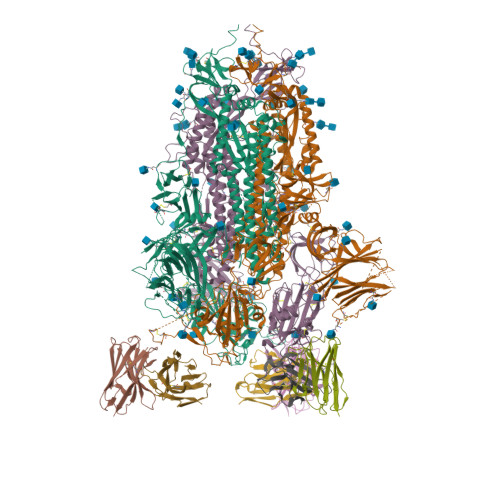
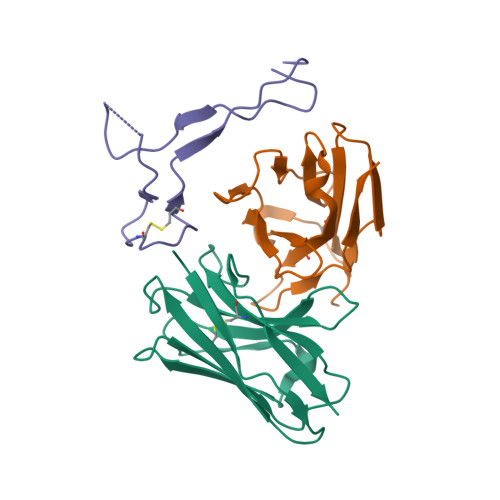
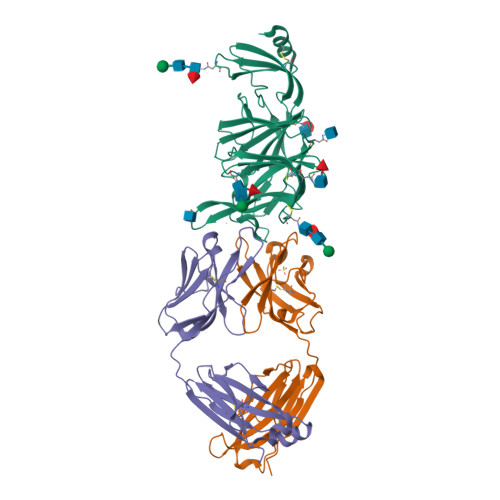
PDB:
6VYB
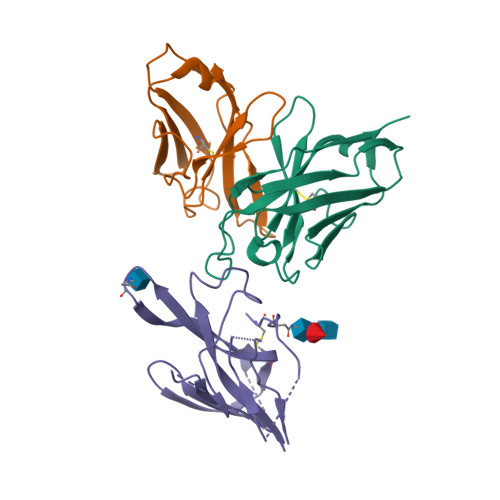
Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology
Wall A, Veesler D, Park YJ, Cameroni E, Lanzavecchia A, Corti D, Sallusto F, Tortorici MA, Pinto D, Beltramello M, Jaconi S, Zatta F, De Marco A, Peter A, Guarino B, Havenar-Daughton C, Snell G, Virgin HW, Fink K, Bowen J, Piccoli L, Czudnochowski N, Silacci-Fregni C, Rosen L, Acton O, Minola A, Sprugasci N, Bassi J, Nix J, Mele F, Jovic S, Rodriguez B, Gupta S, Jin F, Piumatti G, Presti G, Pellanda A, Biggiogero M, Tarkowski M, Pizzuto M, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A
Cell - 2020
volume 183, issue 4, pages 1024-1042.e21
PMID: 32991844; PMCID: PMC7494283
Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms
Ruiz M, Veesler D, Cameroni E, Corti D, Dang HV, Tortorici MA, Pinto D, Beltramello M, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Case JB, Chen R, Havenar-Daughton C, Snell G, Virgin HW, Diamond M, Fink K, McCallum M, Bowen J, Czudnochowski N, Rosen L, Minola A, Sprugasci N, Pizzuto M, Riva A, Lempp F, Lauron E, Tucker H, Zhou J, Wojcechowskyj J, Kaiser H, Meury M, Dillen J, Ng C, Benigni F, Abdelnabi R, Foo SYC, Schmid M, Gabrieli A, Galli M, Neyts J
Science - 2020
volume 370, issue 6519, pages 950-957
PMID: 32972994; PMCID: PMC7857395
N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2
Ruiz M, Walls AC, Veesler D, Tortorici MA, Pinto D, Beltramello M, Bianchi S, Zatta F, De Marco A, Guarino B, McCallum M, Bowen J, Silacci-Fregni C, Rosen L, Lempp F, Zhou J, Abdelnabi R, Foo SYC, Chen A, Liu Z, Zepeda S, di Iulio J, Rothlauf PW
Cell - 2021
volume 184, issue 9, pages 2332-2347
PMID: 33761326; PMCID: PMC7962585
SARS-CoV-2 immune evasion by the variant B.1.427/B.1.429 variant of concern
Van Voorhis WC, Walls AC, Veesler D, Cameroni E, Corti D, Tortorici MA, Pinto D, Bianchi S, Jaconi S, Culap K, De Marco A, Snell G, Telenti A, Virgin HW, McCallum M, Bowen J, Navarro M, Piccoli L, Silacci-Fregni C, Rosen L, Bassi J, Pellanda A, Pizzuto M, Garzoni C, Chen A, di Iulio J, Tilles SW, Saliba Christian, Agostini M, Guastalla SB, Bona G
Science - 2021
volume 373, issue 6555, pages 648-654
PMID: 34210893; PMCID: PMC8020983
Broad sarbecovirus neutralization by a human monoclonal antibody
Ruiz M, Walls AC, Veesler D, Cameroni E, Corti D, Wang Z, Tortorici MA, Pinto D, Beltramello M, Jaconi S, Culap K, Zatta F, De Marco A, Guarino B, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Bowen J, Bloom J, Starr T, Czudnochowski N, Silacci-Fregni C, Rosen L, Sprugasci N, Pizzuto M, Lempp F, Zhou J, Kaiser H, Benigni F, Abdelnabi R, Foo SYC, Schmid M, Neyts J, Liu Z, Zepeda S, di Iulio J, Saliba Christian, Marzi R, Bartha I, Housley MP, Dellota E, Addetia A, Vetti E, Giacchetto-Sasselli I, Whelan SPJ
Nature - 2021
volume 597, pages 103–108
PMID: 34280951; PMCID: PMC9341430
SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape
Veesler D, Park YJ, Cameroni E, Corti D, Pinto D, Beltramello M, Jaconi S, Culap K, Zatta F, De Marco A, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Bowen J, Dingens A, Bloom J, Starr T, Greaney A, Czudnochowski N, Rosen L, Sprugasci N, Nix J, Pizzuto M, Lempp F, Tucker H, Zhou J, Wojcechowskyj J, Kaiser H, Meury M, Liu Z, di Iulio J, Agostini M, Marzi R, Housley MP, Dellota E, Addetia A, Whelan SPJ, Hernandez P, Glass WG, Zhang I, Montiel_Ruiz M, Lombardo G, Croll TI, Chodera JD, Hebner CM
Nature - 2021
volume 597, issue 7874, pages 97-102
PMID: 34261126; PMCID: PMC9282883
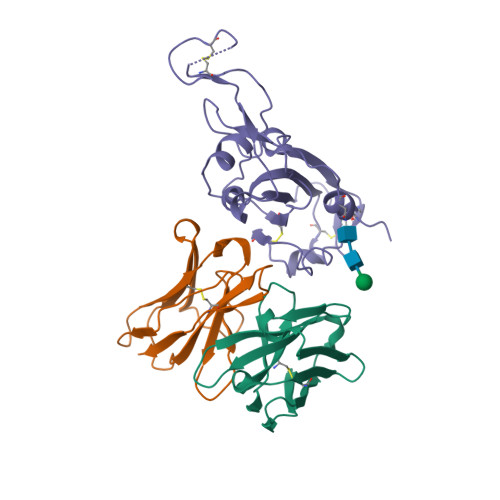
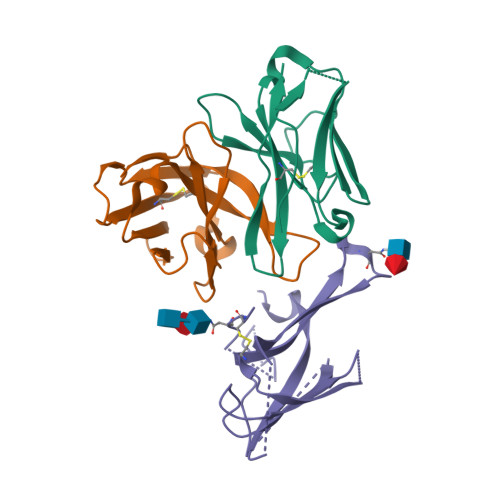
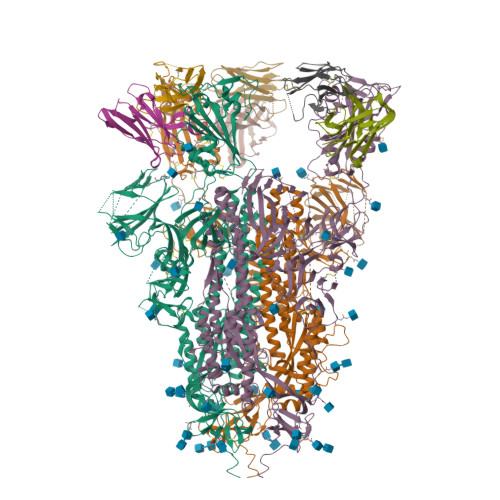
PDB:
7R7N
Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice
Stewart L, Walls AC, Veesler D, Park YJ, Carter L, Ravichandran R, Baker D, Case JB, Chen R, Diamond M, Bowen J, Kozodoy L, Goreshnik I, Cao L, Chow C, Bloom J, Gale M, Starr T, Liu Z, Addetia A, Whelan SPJ, Yeh H, Hunt AC, Wu K, Saini S, Zhao YT, Green LB, Matochko WL, Thomson CA, Voegeli B, Krueger-Gericke A, VanBlargan LA, Ying B, Bailey AL, Kafai NM, Boyken S, Ljubetic A, Edman N, Ueda G, Pranpradist N, Freedman B, Lutz B, Ruohola-Baker H, Jewett MC
Sci Transl Med - 2021
volume 14, issue 646,
PMID: 35412328; PMCID: PMC9258422
Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry
Walls AC, Veesler D, Park YJ, Corti D, Pinto D, Zatta F, De Marco A, Guarino B, Snell G, Virgin HW, Bowen J, Bloom J, Starr T, Pizzuto M, Lempp F, Benigni F, Abdelnabi R, Foo SYC, Neyts J, Liu Z, Zepeda S, Whelan SPJ, Sprouse K, Noack J, Joshi A, Giurdanella M
Science - 2022
volume 375, issue 6579, pages 449-454
PMID: 34990214; PMCID: PMC9400459
Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution
Veesler D, Corti D, Snell G, McCallum M, Bloom J, Loes A, Starr T, Greaney A, Dillen J, Hannon WW, Hauser K, Ferri E, Farrel AG, Dadonaite B, Matreyek KA
Science - 2022
volume 377, issue 6604, pages 420-424
PMID: 35762884; PMCID: PMC9273037
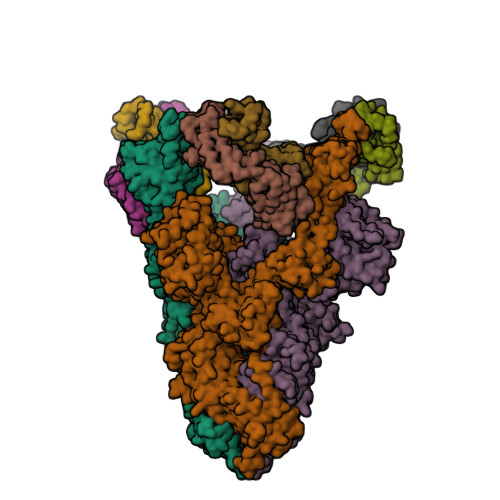
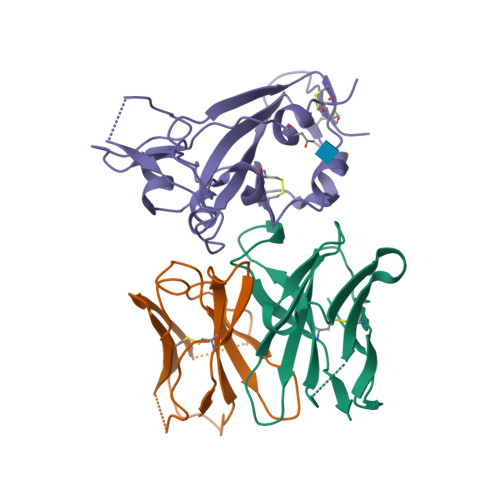
PDB:
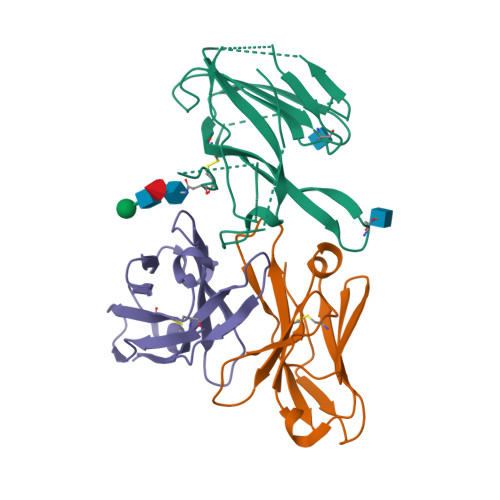
8DF5
ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies
Walls AC, Veesler D, Lanzavecchia A, Corti D, Sallusto F, Tortorici MA, Pinto D, Bianchi S, Snell G, McCallum M, Bowen J, Sprugasci N, Mele F, Ceschi A, Ferrari P, Garzoni C, Benigni F, Abdelnabi R, Neyts J, Low JS, Noack J, Jerak J, Cassotta A, Montiel_Ruiz M, Joshi A, Stewart C, Foglierini M, Weynand B, Rexhepaj M, Jarrossay D, Morone D, Paparoditis P, Purcell LA
Science - 2022
volume 377, issue 6607, pages 735-742
PMID: 35857703; PMCID: PMC9348755
SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines
van Voorhis W, Zhang Z, Tortorici MA, Walls AC, Veesler D, Park YJ, Corti D, Sallusto F, Chu H, Snell G, McCallum M, Bowen J, Sprouse K, Low JS, Jerak J, Franko N, Logue J, Joshi A, Stewart C, Brown JT, Sharkey WK, Mazzitelli IG, Ahmed K, Gori A, Bandera A, Posavad CM, Dan JM, Weiskopf D, Sette A, Crotty S, Iqbal NT, Geffner J, Grifantini R, Nguyen AW, Silva RP, Huang Y, Tiles SW, Shariq A, Maynard JA, Simmerling C
Sci Immunol - 2022
volume 7, issue 78, pages eadf1421
PMID: 36356052; PMCID: PMC9765460
PDB:
8DYA
Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection
Yang F, Walls AC, Veesler D, DiMaio F, Corti D, Dang HV, McCallum M, Kacherovsky N, Cheng E, Cardle I, Sellers D, Salipante S, Pun S
Angew. Chem. Int. Ed. Engl. - 2021
volume 60, issue 39, pages 21211-21215
PMID: 34328683; PMCID: PMC8426805
A broadly generalizable stabilization strategy for sarbecovirus fusion machinery vaccines
Veesler D, Park YJ, Corti D, King NP, Schafer A, Baric R, Sprouse K, Stewart C, Lee J, Leaf EM, Treichel C, Powers J, Asarnow D
Nat Commun - 2024
volume 15, issue 1, pages 5496
PMID: 38944664; PMCID: PMC11254757
A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification
Veesler D, Park YJ, Cameroni E, Lanzavecchia A, Corti D, Dang HV, Tortorici MA, Pinto D, Zatta F, De Marco A, Guarino B, Snell G, Telenti A, Starr T, Czudnochowski N, Rosen L, Lempp F, Benigni F, Liu Z, Zepeda S, di Iulio J, Subramaniam S, Saliba Christian, Dellota E, Addetia A, Whelan SPJ, Sprouse K, Zhang I, Noack J, Giurdanella M, Hauser K, Purcell LA, Acosta R, Rajesh A, Schnell G, de Melo GD, Kergoat L, Bourhy H, Lee J, Taylor AL, Brown J, Foreman WB, Bohan D, Errico JM, Chartron JW, Cusumano G, Tao Q, Schmid MA, Pizzuto MJ
Cell - 2024
volume 187, issue 25, pages 7196-7213
PMID: 39383863; PMCID: PMC11645210
Design of customized coronavirus receptors
Veesler D, Chen Y, Corti D, Yang H, McCallum M, Bowen J, Wang C, Xiong Q, Ma C, Liu C, Si J, Liu P, Shi L, Tong F, Huang M, Li J, Chen J, Yang X, Sun Y, Yu X, Shi Z, Guo H, Guo M
Nature - 2024
volume 635, issue 8040, pages 978-986
PMID: 39478224; PMCID: PMC12187079
External Resources
| RESOURCE | REFERENCE ID |
|---|---|
| BV-BRC: | fig|2697049.107626.CDS.3 |
| GenBank: | QHD43416 |
| UniProt: | P0DTC2 |
Sequences
These sequences are the native gene sequence; sequences of constructs derived from these sequences may differ due to codon optimization or other protocols.
To find the specific sequence of any material you may have ordered, click on the "info" button next to the name of that material.
AA Sequence
MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PRRARSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GAALQIPFAM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TASALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDKVEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQYI KWPWYIWLGF IAGLIAIVMV TIMLCCMTSC CSCLKGCCSC GSCCKFDEDD SEPVLKGVKL HYT
NT Sequence
ATGTTTGTTT TTCTTGTTTT ATTGCCACTA GTCTCTAGTC AGTGTGTTAA TCTTACAACC AGAACTCAAT TACCCCCTGC ATACACTAAT TCTTTCACAC GTGGTGTTTA TTACCCTGAC AAAGTTTTCA GATCCTCAGT TTTACATTCA ACTCAGGACT TGTTCTTACC TTTCTTTTCC AATGTTACTT GGTTCCATGC TATACATGTC TCTGGGACCA ATGGTACTAA GAGGTTTGAT AACCCTGTCC TACCATTTAA TGATGGTGTT TATTTTGCTT CCACTGAGAA GTCTAACATA ATAAGAGGCT GGATTTTTGG TACTACTTTA GATTCGAAGA CCCAGTCCCT ACTTATTGTT AATAACGCTA CTAATGTTGT TATTAAAGTC TGTGAATTTC AATTTTGTAA TGATCCATTT TTGGGTGTTT ATTACCACAA AAACAACAAA AGTTGGATGG AAAGTGAGTT CAGAGTTTAT TCTAGTGCGA ATAATTGCAC TTTTGAATAT GTCTCTCAGC CTTTTCTTAT GGACCTTGAA GGAAAACAGG GTAATTTCAA AAATCTTAGG GAATTTGTGT TTAAGAATAT TGATGGTTAT TTTAAAATAT ATTCTAAGCA CACGCCTATT AATTTAGTGC GTGATCTCCC TCAGGGTTTT TCGGCTTTAG AACCATTGGT AGATTTGCCA ATAGGTATTA ACATCACTAG GTTTCAAACT TTACTTGCTT TACATAGAAG TTATTTGACT CCTGGTGATT CTTCTTCAGG TTGGACAGCT GGTGCTGCAG CTTATTATGT GGGTTATCTT CAACCTAGGA CTTTTCTATT AAAATATAAT GAAAATGGAA CCATTACAGA TGCTGTAGAC TGTGCACTTG ACCCTCTCTC AGAAACAAAG TGTACGTTGA AATCCTTCAC TGTAGAAAAA GGAATCTATC AAACTTCTAA CTTTAGAGTC CAACCAACAG AATCTATTGT TAGATTTCCT AATATTACAA ACTTGTGCCC TTTTGGTGAA GTTTTTAACG CCACCAGATT TGCATCTGTT TATGCTTGGA ACAGGAAGAG AATCAGCAAC TGTGTTGCTG ATTATTCTGT CCTATATAAT TCCGCATCAT TTTCCACTTT TAAGTGTTAT GGAGTGTCTC CTACTAAATT AAATGATCTC TGCTTTACTA ATGTCTATGC AGATTCATTT GTAATTAGAG GTGATGAAGT CAGACAAATC GCTCCAGGGC AAACTGGAAA GATTGCTGAT TATAATTATA AATTACCAGA TGATTTTACA GGCTGCGTTA TAGCTTGGAA TTCTAACAAT CTTGATTCTA AGGTTGGTGG TAATTATAAT TACCTGTATA GATTGTTTAG GAAGTCTAAT CTCAAACCTT TTGAGAGAGA TATTTCAACT GAAATCTATC AGGCCGGTAG CACACCTTGT AATGGTGTTG AAGGTTTTAA TTGTTACTTT CCTTTACAAT CATATGGTTT CCAACCCACT AATGGTGTTG GTTACCAACC ATACAGAGTA GTAGTACTTT CTTTTGAACT TCTACATGCA CCAGCAACTG TTTGTGGACC TAAAAAGTCT ACTAATTTGG TTAAAAACAA ATGTGTCAAT TTCAACTTCA ATGGTTTAAC AGGCACAGGT GTTCTTACTG AGTCTAACAA AAAGTTTCTG CCTTTCCAAC AATTTGGCAG AGACATTGCT GACACTACTG ATGCTGTCCG TGATCCACAG ACACTTGAGA TTCTTGACAT TACACCATGT TCTTTTGGTG GTGTCAGTGT TATAACACCA GGAACAAATA CTTCTAACCA GGTTGCTGTT CTTTATCAGG ATGTTAACTG CACAGAAGTC CCTGTTGCTA TTCATGCAGA TCAACTTACT CCTACTTGGC GTGTTTATTC TACAGGTTCT AATGTTTTTC AAACACGTGC AGGCTGTTTA ATAGGGGCTG AACATGTCAA CAACTCATAT GAGTGTGACA TACCCATTGG TGCAGGTATA TGCGCTAGTT ATCAGACTCA GACTAATTCT CCTCGGCGGG CACGTAGTGT AGCTAGTCAA TCCATCATTG CCTACACTAT GTCACTTGGT GCAGAAAATT CAGTTGCTTA CTCTAATAAC TCTATTGCCA TACCCACAAA TTTTACTATT AGTGTTACCA CAGAAATTCT ACCAGTGTCT ATGACCAAGA CATCAGTAGA TTGTACAATG TACATTTGTG GTGATTCAAC TGAATGCAGC AATCTTTTGT TGCAATATGG CAGTTTTTGT ACACAATTAA ACCGTGCTTT AACTGGAATA GCTGTTGAAC AAGACAAAAA CACCCAAGAA GTTTTTGCAC AAGTCAAACA AATTTACAAA ACACCACCAA TTAAAGATTT TGGTGGTTTT AATTTTTCAC AAATATTACC AGATCCATCA AAACCAAGCA AGAGGTCATT TATTGAAGAT CTACTTTTCA ACAAAGTGAC ACTTGCAGAT GCTGGCTTCA TCAAACAATA TGGTGATTGC CTTGGTGATA TTGCTGCTAG AGACCTCATT TGTGCACAAA AGTTTAACGG CCTTACTGTT TTGCCACCTT TGCTCACAGA TGAAATGATT GCTCAATACA CTTCTGCACT GTTAGCGGGT ACAATCACTT CTGGTTGGAC CTTTGGTGCA GGTGCTGCAT TACAAATACC ATTTGCTATG CAAATGGCTT ATAGGTTTAA TGGTATTGGA GTTACACAGA ATGTTCTCTA TGAGAACCAA AAATTGATTG CCAACCAATT TAATAGTGCT ATTGGCAAAA TTCAAGACTC ACTTTCTTCC ACAGCAAGTG CACTTGGAAA ACTTCAAGAT GTGGTCAACC AAAATGCACA AGCTTTAAAC ACGCTTGTTA AACAACTTAG CTCCAATTTT GGTGCAATTT CAAGTGTTTT AAATGATATC CTTTCACGTC TTGACAAAGT TGAGGCTGAA GTGCAAATTG ATAGGTTGAT CACAGGCAGA CTTCAAAGTT TGCAGACATA TGTGACTCAA CAATTAATTA GAGCTGCAGA AATCAGAGCT TCTGCTAATC TTGCTGCTAC TAAAATGTCA GAGTGTGTAC TTGGACAATC AAAAAGAGTT GATTTTTGTG GAAAGGGCTA TCATCTTATG TCCTTCCCTC AGTCAGCACC TCATGGTGTA GTCTTCTTGC ATGTGACTTA TGTCCCTGCA CAAGAAAAGA ACTTCACAAC TGCTCCTGCC ATTTGTCATG ATGGAAAAGC ACACTTTCCT CGTGAAGGTG TCTTTGTTTC AAATGGCACA CACTGGTTTG TAACACAAAG GAATTTTTAT GAACCACAAA TCATTACTAC AGACAACACA TTTGTGTCTG GTAACTGTGA TGTTGTAATA GGAATTGTCA ACAACACAGT TTATGATCCT TTGCAACCTG AATTAGACTC ATTCAAGGAG GAGTTAGATA AATATTTTAA GAATCATACA TCACCAGATG TTGATTTAGG TGACATCTCT GGCATTAATG CTTCAGTTGT AAACATTCAA AAAGAAATTG ACCGCCTCAA TGAGGTTGCC AAGAATTTAA ATGAATCTCT CATCGATCTC CAAGAACTTG GAAAGTATGA GCAGTATATA AAATGGCCAT GGTACATTTG GCTAGGTTTT ATAGCTGGCT TGATTGCCAT AGTAATGGTG ACAATTATGC TTTGCTGTAT GACCAGTTGC TGTAGTTGTC TCAAGGGCTG TTGTTCTTGT GGATCCTGCT GCAAATTTGA TGAAGACGAC TCTGAGCCAG TGCTCAAAGG AGTCAAATTA CATTACACA