BaboA.01191.a
Dihydrofolate reductase/thymidylate synthase
| CENTER ID: | BaboA.01191.a |
| ORGANISM: | Babesia bovis T2Bo |
| ASSOCIATED DISEASE: | Babesiosis (atypical) |
| CURRENT STATUS: | in PDB |
| COMMUNITY REQUEST: | True |
| NIH RISK GROUP: | 2 |
| SELECT AGENT: | False |
| NIH PRIORITY pathogens category: |
I |
Ordering Clones & Proteins
If there are materials available for this target, they will be listed below.
Materials can be ordered from SSGCID using the button in the "order material" column.
Clicking the button will add the material to a virtual cart.
You may order multiple materials at a time at no cost to you, as this contract is funded
by NIAID. When you are ready to place your order, click the "Place Order" link which will
appear in the top right corner of the page after you place your first item in your cart.
Clones*
| CENTER REFERENCE ID |
DOMAIN/REGION DESCRIPTION |
INFO | AA START |
AA STOP |
ORDER MATERIAL |
|---|---|---|---|---|---|
| BaboA.01191.a.D11.GE00003 | BaboA.01191.a | 1 | 511 | ||
| BaboA.01191.a.D12.GE00001 | BaboA.01191.a | 1 | 188 | ||
| BaboA.01191.a.D13.GE00001 | BaboA.01191.a | 203 | 511 | ||
| BaboA.01191.a.D14.GE00001 | BaboA.01191.a | 203 | 499 | ||
| BaboA.01191.a.D15.GE00001 | BaboA.01191.a | 225 | 511 | ||
| BaboA.01191.a.D16.GE00001 | BaboA.01191.a | 225 | 499 |
* SSGCID clones represent un-induced expression constructs which have been verified by sequencing from vector primers. Clones may contain a tag, such as N-term 6xHis. Get sequence information
using the button in the "info" column.
Proteins
| CENTER REFERENCE ID |
DOMAIN/REGION DESCRIPTION |
INFO | AA START |
AA STOP |
ORDER MATERIAL |
|---|---|---|---|---|---|
| BaboA.01191.a.D13.PD00068 | BaboA.01191.a | 203 | 511 |
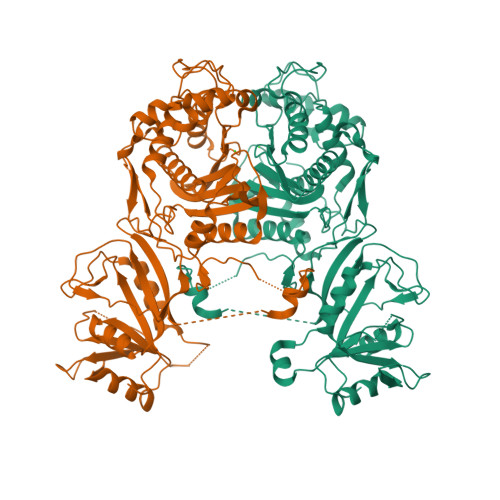
Structures
3I3R
DEPOSITED: 6/30/2009
DETERMINATION: XRay
CLONE: BaboA.01191.a.D11.GE00003
PROTEIN: BaboA.01191.a.D11.PD00043
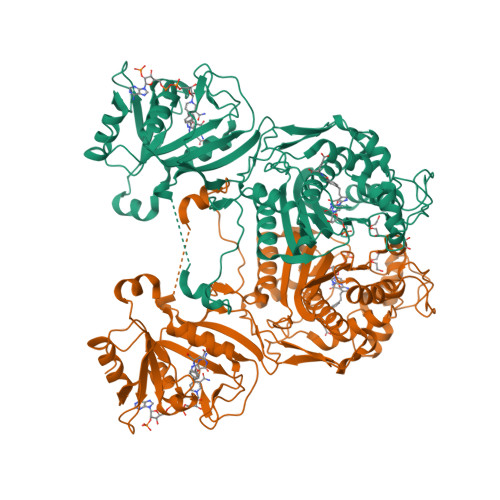
3K2H
DEPOSITED: 9/30/2009
DETERMINATION: XRay
CLONE: BaboA.01191.a.D11.GE00003
PROTEIN: BaboA.01191.a.D11.PD00043
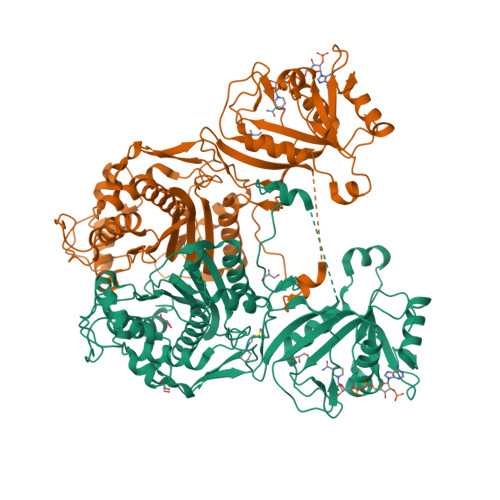
3KJR
DEPOSITED: 11/3/2009
DETERMINATION: XRay
CLONE: BaboA.01191.a.D11.GE00003
PROTEIN: BaboA.01191.a.D11.PD00043
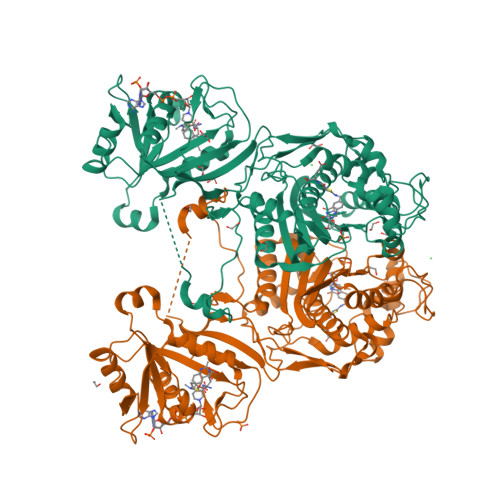
3NRR
DEPOSITED: 6/30/2010
DETERMINATION: XRay
CLONE: BaboA.01191.a.D11.GE00003
PROTEIN: BaboA.01191.a.D11.PD00043
Publications by SSGCID
Inhibitor-bound complexes of dihydrofolate reductase-thymidylate synthase from Babesia bovis
Abendroth J, Begley DW, Edwards TE, Hartley RC, Lorimer DD, Myler PJ, Raymond AC, Sankaran B, Smith ER, Staker BL, Stewart LJ
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. - 2011
volume 67, issue Pt 9, pages 1070-7
PMID: 21904052; PMCID: PMC3169404
External Resources
| RESOURCE | REFERENCE ID |
|---|---|
| EuPathDB: | PiroplasmaDB:BBOV_II000780 |
| RefSeq: | XP_001609606.1 |
| UniProt: | A7ASX7 |
Sequences
These sequences are the native gene sequence; sequences of constructs derived from these sequences may differ due to codon optimization or other protocols.
To find the specific sequence of any material you may have ordered, click on the "info" button next to the name of that material.
AA Sequence
MSNSYEGCGD LTIFVAVALN KVIGHKNQIP WPHITHDFRF LRNGTTYIPP EVLSKNPDIQ NVVIFGRKTY ESIPKASLPL KNRINVILSR TVKEVPGCLV YEDLSTAIRD LRANVPHNKI FILGGSFLYK EVLDNGLCDK IYLTRLNKEY PGDTYFPDIP DTFEITAISP TFSTDFVSYD FVIYERKDCK TVFPDPPFDQ LLMTGTDISV PKPKYVACPG VRIRNHEEFQ YLDILADVLS HGVLKPNRTG TDAYSKFGYQ MRFDLSRSFP LLTTKKVALR SIIEELLWFI KGSTNGNDLL AKNVRIWELN GRRDFLDKNG FTDREEHDLG PIYGFQWRHF GAEYLDMHAD YTGKGIDQLA EIINRIKTNP NDRRLIVCSW NVSDLKKMAL PPCHCFFQFY VSDNKLSCMM HQRSCDLGLG VPFNIASYSI LTAMVAQVCG LGLGEFVHNL ADAHIYVDHV DAVTTQIARI PHPFPRLRLN PDIRNIEDFT IDDIVVEDYV SHPPIPMAMS A
NT Sequence
atgtctaatt catacgaagg gtgtggagat cttacgattt tcgtagccgt tgccttgaat aaggtaattg gccataaaaa ccaaattccc tggccgcata ttacgcatga ttttcgtttt ttgcgtaatg gcacaactta cattccacct gaggtgttgt ctaagaaccc agatatacag aatgttgtga ttttcggtag gaagacttat gaaagtatcc ctaaagcttc gctccctctt aaaaaccgta taaacgtgat actgtctcgt actgtcaagg aagttcctgg ttgtttggtg tacgaggacc tttctactgc cattcgtgat ttgcgtgcaa atgtccctca taacaagatt tttatcttag gcggttcttt tctttacaag gaggtgcttg acaatggttt gtgtgataag atttatttga ctcgtctcaa caaggagtat cctggtgaca cctacttccc tgatattccg gatacttttg agataaccgc cattagtcct acgttttcca ctgactttgt cagttatgat tttgttattt acgagcgtaa ggattgcaag actgttttcc ccgatcctcc ttttgatcaa ttattgatga cagggactga cattagcgtt cccaaaccaa agtacgttgc ttgtcctggc gtcaggatac gcaaccatga agagttccaa taccttgaca tattggcaga tgtgctaagt catggtgtat tgaagcccaa ccgtaccggt actgatgcct actctaagtt tggctatcag atgcgatttg acctttcacg ttcattcccg ttattgacta ccaagaaggt agcattgcgc agcataattg aggaactgct ctggtttatt aagggtagta ccaacggcaa cgacctactg gctaagaatg tgcgcatttg ggagcttaac ggtcgtcgtg atttccttga taagaatggt tttacagatc gtgaagagca tgatttaggt cctatatatg gtttccaatg gcgtcatttt ggtgctgagt acttggatat gcacgctgat tacactggta agggtattga tcagcttgct gagataatta accgcattaa aacgaaccca aacgaccgta ggctgattgt atgctcttgg aatgtttcgg acctaaagaa gatggctttg cctccttgtc attgtttctt ccaattttat gtcagtgaca acaagctttc ttgtatgatg catcagcgtt cctgtgacct tggtttgggt gtacctttca acattgcttc ttatagtata cttactgcta tggtggctca ggtttgcggt ttaggtttag gggagtttgt ccataatttg gcagacgccc acatttatgt tgaccacgtg gacgccgtta ctactcagat tgctcgcatt cctcatccat ttccccgttt gcgtttgaac cctgatatta ggaatatcga ggactttact attgacgaca ttgtagttga ggattacgtc tcgcatcctc ccatacccat ggccatgtcc gcatga
Details for BaboA.01191.a.D11.GE00003
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Moderate Expression |
| SOLUBLE EXPRESSION LEVEL | Moderate Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass with incomplete coverage |
| PERCENT IDENTITY: | 81 |
| PERCENT COVERAGE: | 63 |
Validated AA Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMSNS YEGCGDLTIF VAVALNKVIG HKNQIPWPHI THDFRFLRNG TTYIPPEVLS KNPDIQNVVI FGRKTYESIP KASLPLKNRI NVILSRTVKE VPGCLVYEDL STAIRDLRAN VPHNKIFILG GSFLYKEVLD NGLCDKIYLT RLNKEYPGDT YFPDIPDTFE ITAISPTFST DFVSYDFVIY ERKDCKTVFP DPPFXXLLMT GTDISVPKPK YVXXPXFVXV TXKNXXIXTS XLXXCPXXCX XXXXXXXLXX XXXXXXXXXX XLXXXXXXXX XXXXXXXXXX XXXXXX
Validated NT Sequence
ctnnnnnnat tttgtttaac tttaagaagg agatatacca tgggccacca ccaccaccac catagcggcg aagtaaaacc ggaagtgaag ccggagaccc acatcaacct gaaggttagc gacggtagca gcgaaatctt cttcaagatt aagaagacca ccccgctgcg tcgcctgatg gaagcattcg ctaaacgcca gggcaaggaa atggattccc tgcgctttct gtacgacggt atccgtattc aggcagacca gactccggaa gatctggaca tggaagataa cgacattatc gaagcacacc gtgaacaaat cggtggaatg tccaatagct atgaaggttg tggcgatctg actatcttcg tagctgtcgc actcaacaaa gttatcggtc ataaaaatca gatcccttgg ccacacataa cccatgactt tcgtttcctg cgtaacggca ccacctacat cccgccggaa gtactgagca agaatcctga tatccagaac gtggtgatct tcggccgtaa aacttacgag tctatcccta aggcctcgct gccgctcaag aatcgtatca acgtaatcct ctctcgtacc gttaaggaag tgcctggttg cttagtttat gaagatctgt ccactgcgat tcgcgacctg cgtgcaaacg tacctcacaa caagatcttc atcctgggcg gttccttcct ctacaaagaa gttcttgaca acggtctgtg tgacaagatc tatctgaccc gcctcaacaa ggaatacccg ggcgacactt acttcccgga tatccctgat accttcgaaa taaccgccat ctctcctacc ttctccactg acttcgtttc ttacgatttc gtaatttacg agcgtaaaga ttgcaagact gtgttcccgg acccgccatt tgancnactc ctgatgaccg gcaccgacat ctctgttcct aaaccgaagt acgtancctg nccggngttc gtatncgtaa ccncnaagaa tnnnagnatc ntgacntctn agctgannng ctgtccnnng nngtgctnan ncnancnncn ncngnnnnna ctnnnnnnaa nttnnnnncn nnngnnnnnn nnnnnncnnn anctncnncn cnnncnnnan annnnnnnnn nnnnnnnnnn nnnnnncnnn nnncnnnnnn nnnnnnnnnn nnnnnnaa
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMSNS YEGCGDLTIF VAVALNKVIG HKNQIPWPHI THDFRFLRNG TTYIPPEVLS KNPDIQNVVI FGRKTYESIP KASLPLKNRI NVILSRTVKE VPGCLVYEDL STAIRDLRAN VPHNKIFILG GSFLYKEVLD NGLCDKIYLT RLNKEYPGDT YFPDIPDTFE ITAISPTFST DFVSYDFVIY ERKDCKTVFP DPPFDQLLMT GTDISVPKPK YVACPGVRIR NHEEFQYLDI LADVLSHGVL KPNRTGTDAY SKFGYQMRFD LSRSFPLLTT KKVALRSIIE ELLWFIKGST NGNDLLAKNV RIWELNGRRD FLDKNGFTDR EEHDLGPIYG FQWRHFGAEY LDMHADYTGK GIDQLAEIIN RIKTNPNDRR LIVCSWNVSD LKKMALPPCH CFFQFYVSDN KLSCMMHQRS CDLGLGVPFN IASYSILTAM VAQVCGLGLG EFVHNLADAH IYVDHVDAVT TQIARIPHPF PRLRLNPDIR NIEDFTIDDI VVEDYVSHPP IPMAMSA
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaat gtccaatagc tatgaaggtt gtggcgatct gactatcttc gtagctgtcg cactcaacaa agttatcggt cataaaaatc agatcccttg gccacacata acccatgact ttcgtttcct gcgtaacggc accacctaca tcccgccgga agtactgagc aagaatcctg atatccagaa cgtggtgatc ttcggccgta aaacttacga gtctatccct aaggcctcgc tgccgctcaa gaatcgtatc aacgtaatcc tctctcgtac cgttaaggaa gtgcctggtt gcttagttta tgaagatctg tccactgcga ttcgcgacct gcgtgcaaac gtacctcaca acaagatctt catcctgggc ggttccttcc tctacaaaga agttcttgac aacggtctgt gtgacaagat ctatctgacc cgcctcaaca aggaataccc gggcgacact tacttcccgg atatccctga taccttcgaa ataaccgcca tctctcctac cttctccact gacttcgttt cttacgattt cgtaatttac gagcgtaaag attgcaagac tgtgttcccg gacccgccat ttgaccaact cctgatgacc ggcaccgaca tctctgttcc taaaccgaag tacgtagcct gccctggcgt tcgtatccgt aaccacgaag aattccagta tcttgacatc ttagctgacg tgctgtccca cggtgtgctg aagccgaacc gcaccggcac tgacgcttac tcgaaatttg gttaccagat gcgcttcgac ctttcccgta gcttcccgct cctgaccact aagaaggttg cgctccgctc tatcatcgag gaactgctct ggtttatcaa aggttctacc aacggtaacg atctgttagc taagaatgtt cgtatctggg aactgaacgg tcgtcgtgac tttcttgaca agaatggttt cactgatcgc gaagagcacg acctgggtcc gatttacggc ttccagtggc gtcactttgg tgctgaatac cttgacatgc acgctgatta cactggcaaa ggtatcgatc agttagctga aatcattaat cgcatcaaga ctaacccgaa cgatcgccgt ctgatcgttt gctcctggaa cgtctctgat ctcaagaaaa tggcactgcc gccttgtcac tgcttcttcc agttctatgt tagcgacaac aaactgagct gcatgatgca ccagcgtagc tgcgatctgg gcctgggcgt tccgttcaat atcgcttcct actccattct gaccgctatg gttgcccaag tttgtggtct tggtctgggt gaattcgttc ataacctggc tgatgcccac atttacgtgg accacgttga cgctgtgacc acccagatag ctcgtattcc tcacccattt ccgcgcctgc gtcttaatcc agacattcgt aacattgaag acttcaccat cgacgatatc gtcgtggaag attacgtgtc ccacccaccg attcctatgg ccatgtctgc ataaaagctt gcggccgcac tcgagcacca ccaccaccac cactgagatc cggctgctaa caaagcccga aaggaagctg agttggctgc tgccaccgct gagcaataac tagcataacc ccttggggcc tctaaacggg tcttgagggg ttttttgctg aaaggaggaa ctatatccgg at
Details for BaboA.01191.a.D12.GE00001
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Low Expression |
| SOLUBLE EXPRESSION LEVEL | Low Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 100 |
Validated AA Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMSNS YEGCGDLTIF VAVALNKVIG HKNQIPWPHI THDFRFLRNG TTYIPPEVLS KNPDIQNVVI FGRKTYESIP KASLPLKNRI NVILSRTVKE VPGCLVYEDL STAIRDLRAN VPHNKIFILG GSFLYKEVLD NGLCDKIYLT RLNKEYPGDT YFPDIPDTFE ITAISPTFST DFVSYDFVIY ERKD
Validated NT Sequence
atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaat gtccaatagc tatgaaggtt gtggcgatct gactatcttc gtagctgtcg cactcaacaa agttatcggt cataaaaatc agatcccttg gccacacata acccatgact ttcgtttcct gcgtaacggc accacctaca tcccgccgga agtactgagc aagaatcctg atatccagaa cgtggtgatc ttcggccgta aaacttacga gtctatccct aaggcctcgc tgccgctcaa gaatcgtatc aacgtaatcc tctctcgtac cgttaaggaa gtgcctggtt gcttagttta tgaagatctg tccactgcga ttcgcgacct gcgtgcaaac gtacctcaca acaagatctt catcctgggc ggttccttcc tctacaaaga agttcttgac aacggtctgt gtgacaagat ctatctgacc cgcctcaaca aggaataccc gggcgacact tacttcccgg atatccctga taccttcgaa ataaccgcca tctctcctac cttctccact gacttcgttt cttacgattt cgtaatttac gagcgtaaag attaa
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMSNS YEGCGDLTIF VAVALNKVIG HKNQIPWPHI THDFRFLRNG TTYIPPEVLS KNPDIQNVVI FGRKTYESIP KASLPLKNRI NVILSRTVKE VPGCLVYEDL STAIRDLRAN VPHNKIFILG GSFLYKEVLD NGLCDKIYLT RLNKEYPGDT YFPDIPDTFE ITAISPTFST DFVSYDFVIY ERKD
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaat gtccaatagc tatgaaggtt gtggcgatct gactatcttc gtagctgtcg cactcaacaa agttatcggt cataaaaatc agatcccttg gccacacata acccatgact ttcgtttcct gcgtaacggc accacctaca tcccgccgga agtactgagc aagaatcctg atatccagaa cgtggtgatc ttcggccgta aaacttacga gtctatccct aaggcctcgc tgccgctcaa gaatcgtatc aacgtaatcc tctctcgtac cgttaaggaa gtgcctggtt gcttagttta tgaagatctg tccactgcga ttcgcgacct gcgtgcaaac gtacctcaca acaagatctt catcctgggc ggttccttcc tctacaaaga agttcttgac aacggtctgt gtgacaagat ctatctgacc cgcctcaaca aggaataccc gggcgacact tacttcccgg atatccctga taccttcgaa ataaccgcca tctctcctac cttctccact gacttcgttt cttacgattt cgtaatttac gagcgtaaag attaaaagct tgcggccgca ctcgagcacc accaccacca ccactgagat ccggctgcta acaaagcccg aaaggaagct gagttggctg ctgccaccgc tgagcaataa ctagcataac cccttggggc ctctaaacgg gtcttgaggg gttttttgct gaaaggagga actatatccg gat
Details for BaboA.01191.a.D13.GE00001
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Moderate Expression |
| SOLUBLE EXPRESSION LEVEL | Moderate Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 61 |
Validated AA Sequence
MRFDLSRSFP LLTTKKVALR SIIEELLWFI KGSTNGNDLL AKNVRIWELN GRRDFLDKNG FTDREEHDLG PIYGFQWRHF GAEYLDMHAD YTGKGIDQLA EIINRIKTNP NDRRLIVCSW NVSDLKKMAL PPCHCFFQFY VSDNKLSCMM HQRSCDLGLG VPFNIASYSI LTAMVAQVCG LGLGEFVHNL ADAHIYVDHV DAVTTQIARI PHPFPRLRLN PDIRNIEDFT IDDIVVEDYV SHPPIPMAMS A
Validated NT Sequence
ttcnggcttt gttagcagcc ggntctcngt ggtggtggtg gtggtgctcg agtgcggccg caagctttta tgcagacatg gccataggaa tcggtgggtg ggacacgtaa tcttccacga cgatatcgtc gatggtgaag tcttcaatgt tacgaatgtc tggattaaga cgcaggcgcg gaaatgggtg aggaatacga gctatctggg tggtcacagc gtcaacgtgg tccacgtaaa tgtgggcatc agccaggtta tgaacgaatt cacccagacc aagaccacaa acttgggcaa ccatagcggt cagaatggag taggaagcga tattgaacgg aacgcccagg cccagatcgc agctacgctg gtgcatcatg cagctcagtt tgttgtcgct aacatagaac tggaagaagc agtgacaagg cggcagtgcc attttcttga gatcagagac gttccaggag caaacgatca gacggcgatc gttcgggtta gtcttgatgc gattaatgat ttcagctaac tgatcgatac ctttgccagt gtaatcagcg tgcatgtcaa ggtattcagc accaaagtga cgccactgga agccgtaaat cggacccagg tcgtgctctt cgcgatcagt gaaaccattc ttgtcaagaa agtcacgacg accgttcagt tcccagatac gaacattctt agctaacaga tcgttaccgt tggtagaacc tttgataaac cagagcagtt cctcgatgat agagcggagc gcaaccttct tagtggtcag gagcgggaag ctacgggaaa ggtcgaagcg catctggtaa ccaaatttcg agtaagcgtc agtgccngtg cggttcggct tcagcacacc gtgggacagc acgtcagcta agatgtcnag atactggaat tcttcgtggt tnnngatacg aacgccnggn nggctacgta cttcggtttn nnaacagann ngtcggtgnc gntcattcca ccgattnnnn cangnnnnnc ntcgannatg tcntnntcnt nnntgtncag atnntnnnnn nnnngnctnn cnnantangn nnnccnnnnn nnaaagcnnn nnnntnnnnt tnnnnnnncn nnnnntnnnn nanngnnncn nnngnnnnnn nnnnngnnnn nnnnnnnnnn nnanancnnn nnnnnnnnnn gnnnnaannn nnnnnnnccg g
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMTGT DISVPKPKYV ACPGVRIRNH EEFQYLDILA DVLSHGVLKP NRTGTDAYSK FGYQMRFDLS RSFPLLTTKK VALRSIIEEL LWFIKGSTNG NDLLAKNVRI WELNGRRDFL DKNGFTDREE HDLGPIYGFQ WRHFGAEYLD MHADYTGKGI DQLAEIINRI KTNPNDRRLI VCSWNVSDLK KMALPPCHCF FQFYVSDNKL SCMMHQRSCD LGLGVPFNIA SYSILTAMVA QVCGLGLGEF VHNLADAHIY VDHVDAVTTQ IARIPHPFPR LRLNPDIRNI EDFTIDDIVV EDYVSHPPIP MAMSA
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaat gaccggcacc gacatctctg ttcctaaacc gaagtacgta gcctgccctg gcgttcgtat ccgtaaccac gaagaattcc agtatcttga catcttagct gacgtgctgt cccacggtgt gctgaagccg aaccgcaccg gcactgacgc ttactcgaaa tttggttacc agatgcgctt cgacctttcc cgtagcttcc cgctcctgac cactaagaag gttgcgctcc gctctatcat cgaggaactg ctctggttta tcaaaggttc taccaacggt aacgatctgt tagctaagaa tgttcgtatc tgggaactga acggtcgtcg tgactttctt gacaagaatg gtttcactga tcgcgaagag cacgacctgg gtccgattta cggcttccag tggcgtcact ttggtgctga ataccttgac atgcacgctg attacactgg caaaggtatc gatcagttag ctgaaatcat taatcgcatc aagactaacc cgaacgatcg ccgtctgatc gtttgctcct ggaacgtctc tgatctcaag aaaatggcac tgccgccttg tcactgcttc ttccagttct atgttagcga caacaaactg agctgcatga tgcaccagcg tagctgcgat ctgggcctgg gcgttccgtt caatatcgct tcctactcca ttctgaccgc tatggttgcc caagtttgtg gtcttggtct gggtgaattc gttcataacc tggctgatgc ccacatttac gtggaccacg ttgacgctgt gaccacccag atagctcgta ttcctcaccc atttccgcgc ctgcgtctta atccagacat tcgtaacatt gaagacttca ccatcgacga tatcgtcgtg gaagattacg tgtcccaccc accgattcct atggccatgt ctgcataaaa gcttgcggcc gcactcgagc accaccacca ccaccactga gatccggctg ctaacaaagc ccgaaaggaa gctgagttgg ctgctgccac cgctgagcaa taactagcat aaccccttgg ggcctctaaa cgggtcttga ggggtttttt gctgaaagga ggaactatat ccggat
Details for BaboA.01191.a.D14.GE00001
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Moderate Expression |
| SOLUBLE EXPRESSION LEVEL | No Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 88 |
Validated AA Sequence
MEAFAKRQGK EMDSLRFLYD GIRIQADQTP EDLDMEDNDI IEAHREQIGG MTGTDISVPK PKYVACPGVR IRNHEEFQYL DILADVLSHG VLKPNRTGTD AYSKFGYQMR FDLSRSFPLL TTKKVALRSI IEELLWFIKG STNGNDLLAK NVRIWELNGR RDFLDKNGFT DREEHDLGPI YGFQWRHFGA EYLDMHADYT GKGIDQLAEI INRIKTNPND RRLIVCSWNV SDLKKMALPP CHCFFQFYVS DNKLSCMMHQ RSCDLGLGVP FNIASYSILT AMVAQVCGLG LGEFVHNLAD AHIYVDHVDA VTTQIARIPH PFPRLRLNPD IRNIEDFTID DIVVEDY
Validated NT Sequence
tagcggcgaa gtaaaaccgg aagtgaagcc ggagacccac atcaacctga aggttagcga cggtagcagc gaaatcttct tcaagattaa gaagaccacc ccgctgcgtc gcctgatgga agcattcgct aaacgccagg gcaaggaaat ggattccctg cgctttctgt acgacggtat ccgtattcag gcagaccaga ctccggaaga tctggacatg gaagataacg acattatcga agcacaccgt gaacaaatcg gtggaatgac cggcaccgac atctctgttc ctaaaccgaa gtacgtagcc tgccctggcg ttcgtatccg taaccacgaa gaattccagt atcttgacat cttagctgac gtgctgtccc acggtgtgct gaagccgaac cgcaccggca ctgacgctta ctcgaaattt ggttaccaga tgcgcttcga cctttcccgt agcttcccgc tcctgaccac taagaaggtt gcgctccgct ctatcatcga ggaactgctc tggtttatca aaggttctac caacggtaac gatctgttag ctaagaatgt tcgtatctgg gaactgaacg gtcgtcgtga ctttcttgac aagaatggtt tcactgatcg cgaagagcac gacctgggtc cgatttacgg cttccagtgg cgtcactttg gtgctgaata ccttgacatg cacgctgatt acactggcaa aggtatcgat cagttagctg aaatcattaa tcgcatcaag actaacccga acgatcgccg tctgatcgtt tgctcctgga acgtctctga tctcaagaaa atggcactgc cgccttgtca ctgcttcttc cagttctatg ttagcgacaa caaactgagc tgcatgatgc accagcgtag ctgcgatctg ggcctgggcg ttccgttcaa tatcgcttcc tactccattc tgaccgctat ggttgcccaa gtttgtggtc ttggtctggg tgaattcgtt cataacctgg ctgatgccca catttacgtg gaccacgttg acgctgtgac cacccagata gctcgtattc ctcacccatt tccgcgcctg cgtcttaatc cagacattcg taacattgaa gacttcacca tcgacgatat cgtcgtggaa gattacta
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMTGT DISVPKPKYV ACPGVRIRNH EEFQYLDILA DVLSHGVLKP NRTGTDAYSK FGYQMRFDLS RSFPLLTTKK VALRSIIEEL LWFIKGSTNG NDLLAKNVRI WELNGRRDFL DKNGFTDREE HDLGPIYGFQ WRHFGAEYLD MHADYTGKGI DQLAEIINRI KTNPNDRRLI VCSWNVSDLK KMALPPCHCF FQFYVSDNKL SCMMHQRSCD LGLGVPFNIA SYSILTAMVA QVCGLGLGEF VHNLADAHIY VDHVDAVTTQ IARIPHPFPR LRLNPDIRNI EDFTIDDIVV EDY
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaat gaccggcacc gacatctctg ttcctaaacc gaagtacgta gcctgccctg gcgttcgtat ccgtaaccac gaagaattcc agtatcttga catcttagct gacgtgctgt cccacggtgt gctgaagccg aaccgcaccg gcactgacgc ttactcgaaa tttggttacc agatgcgctt cgacctttcc cgtagcttcc cgctcctgac cactaagaag gttgcgctcc gctctatcat cgaggaactg ctctggttta tcaaaggttc taccaacggt aacgatctgt tagctaagaa tgttcgtatc tgggaactga acggtcgtcg tgactttctt gacaagaatg gtttcactga tcgcgaagag cacgacctgg gtccgattta cggcttccag tggcgtcact ttggtgctga ataccttgac atgcacgctg attacactgg caaaggtatc gatcagttag ctgaaatcat taatcgcatc aagactaacc cgaacgatcg ccgtctgatc gtttgctcct ggaacgtctc tgatctcaag aaaatggcac tgccgccttg tcactgcttc ttccagttct atgttagcga caacaaactg agctgcatga tgcaccagcg tagctgcgat ctgggcctgg gcgttccgtt caatatcgct tcctactcca ttctgaccgc tatggttgcc caagtttgtg gtcttggtct gggtgaattc gttcataacc tggctgatgc ccacatttac gtggaccacg ttgacgctgt gaccacccag atagctcgta ttcctcaccc atttccgcgc ctgcgtctta atccagacat tcgtaacatt gaagacttca ccatcgacga tatcgtcgtg gaagattact aaaagcttgc ggccgcactc gagcaccacc accaccacca ctgagatccg gctgctaaca aagcccgaaa ggaagctgag ttggctgctg ccaccgctga gcaataacta gcataacccc ttggggcctc taaacgggtc ttgaggggtt ttttgctgaa aggaggaact atatccggat
Details for BaboA.01191.a.D15.GE00001
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Moderate Expression |
| SOLUBLE EXPRESSION LEVEL | No Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass |
| PERCENT IDENTITY: | 98 |
| PERCENT COVERAGE: | 79 |
Validated AA Sequence
MXDNDXXEAX XEQIGGNHEE FQYLDILADV LSHGVLKPNR TGTDAYSKFG YQMRFDLSXS FPLLTTKKVA LRSIIEELLW FIKGSTNGND LLAKNVRIWE LNGRRDFLDK NGFTDREEHD LGPIYGFQWR HFGAEYLDMH ADYTGKGIDQ LAEIINRIKT NPNDRRLIVC SWNVSDLKKM ALPPCHCFFQ FYVSDNKLSC MMHQRSCDLG LGVPFNIASY SILTAMVAQV CGLGLGEFVH NLADAHIYVD HVDAVTTQIA RIPHPFPRLR LNPDIRNIED FTIDDIVVED YVSHPPIPMA MSA
Validated NT Sequence
tnntncnggc tttgttagca gccggnnctc agtggtggtg gtggtggtgc tcgagtgcgg ccgcaagctt ttatgcagac atggccatag gaatcggtgg gtgggacacg taatcttcca cgacgatatc gtcgatggtg aagtcttcaa tgttacgaat gtctggatta agacgcaggc gcggaaatgg gtgaggaata cgagctatct gggtggtcac agcgtcaacg tggtccacgt aaatgtgggc atcagccagg ttatgaacga attcacccag accaagacca caaacttggg caaccatagc ggtcagaatg gagtaggaag cgatattgaa cggaacgccc aggcccagat cgcagctacg ctggtgcatc atgcagctca gtttgttgtc gctaacatag aactggaaga agcagtgaca aggcggcagt gccattttct tgagatcaga gacgttccag gagcaaacga tcagacggcg atcgttcggg ttagtcttga tgcgattaat gatttcagct aactgatcga tacctttgcc agtgtaatca gcgtgcatgt caaggtattc agcaccaaag tgacgccact ggaagccgta aatcggaccc aggtcgtgct cttcgcgatc agtgaaacca ttcttgtcaa gaaagtcacg acgaccgttc agttcccaga tacgaacatt cttagctaac agatcgttac cgttggtaga acctttgata aaccagagca gttcctcgat gatagagcgg agcgcaacct tcttagtggt caggagcggg aagctacngg aaaggtcgaa gcgcatctgg taaccaaatt tcgagtaagc gtcagtgccg gtgcggttcg gcttcagcac accgtgggac agcacgtcag ctaagatgtc aagatactgg aattcttcgt ggtttccacc gatttgttca ngntgtgctt cnntantgtc gttatcttnc atgtnnagat cttncnnant ctggtctnnn tgnanannna tancgncnna canaaagcgc nnnnnnnatt tnnnnnccnn gnntnnncna ntgcttcnnc ngnnannggg nnnnnnnnnn nnnnnnannn tnncnnctnc nnnnctnann nnnnnnnnnn nntnnnnnnn nnnnnnnnnn nnnnncc
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGNHEE FQYLDILADV LSHGVLKPNR TGTDAYSKFG YQMRFDLSRS FPLLTTKKVA LRSIIEELLW FIKGSTNGND LLAKNVRIWE LNGRRDFLDK NGFTDREEHD LGPIYGFQWR HFGAEYLDMH ADYTGKGIDQ LAEIINRIKT NPNDRRLIVC SWNVSDLKKM ALPPCHCFFQ FYVSDNKLSC MMHQRSCDLG LGVPFNIASY SILTAMVAQV CGLGLGEFVH NLADAHIYVD HVDAVTTQIA RIPHPFPRLR LNPDIRNIED FTIDDIVVED YVSHPPIPMA MSA
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaaa ccacgaagaa ttccagtatc ttgacatctt agctgacgtg ctgtcccacg gtgtgctgaa gccgaaccgc accggcactg acgcttactc gaaatttggt taccagatgc gcttcgacct ttcccgtagc ttcccgctcc tgaccactaa gaaggttgcg ctccgctcta tcatcgagga actgctctgg tttatcaaag gttctaccaa cggtaacgat ctgttagcta agaatgttcg tatctgggaa ctgaacggtc gtcgtgactt tcttgacaag aatggtttca ctgatcgcga agagcacgac ctgggtccga tttacggctt ccagtggcgt cactttggtg ctgaatacct tgacatgcac gctgattaca ctggcaaagg tatcgatcag ttagctgaaa tcattaatcg catcaagact aacccgaacg atcgccgtct gatcgtttgc tcctggaacg tctctgatct caagaaaatg gcactgccgc cttgtcactg cttcttccag ttctatgtta gcgacaacaa actgagctgc atgatgcacc agcgtagctg cgatctgggc ctgggcgttc cgttcaatat cgcttcctac tccattctga ccgctatggt tgcccaagtt tgtggtcttg gtctgggtga attcgttcat aacctggctg atgcccacat ttacgtggac cacgttgacg ctgtgaccac ccagatagct cgtattcctc acccatttcc gcgcctgcgt cttaatccag acattcgtaa cattgaagac ttcaccatcg acgatatcgt cgtggaagat tacgtgtccc acccaccgat tcctatggcc atgtctgcat aaaagcttgc ggccgcactc gagcaccacc accaccacca ctgagatccg gctgctaaca aagcccgaaa ggaagctgag ttggctgctg ccaccgctga gcaataacta gcataacccc ttggggcctc taaacgggtc ttgaggggtt ttttgctgaa aggaggaact atatccggat
Details for BaboA.01191.a.D16.GE00001
| HARVESTED ON: | data unavailable |
| SEQUENCED ON: | 7/28/2009 |
| EXPECTED MW: | 0kDa |
| OBSERVED MW: | 0kDa |
| ANTIBIOTIC MARKER: | kanamycin |
| COUNT OF EXPRESSION COLONIES: | data unavailable |
| TOTAL EXPRESSION LEVEL: | Moderate Expression |
| SOLUBLE EXPRESSION LEVEL | No Expression |
| EXPRESSION HOST: | data unavailable |
| SEQUENCING RESULT: | pass |
| PERCENT IDENTITY: | 95 |
| PERCENT COVERAGE: | 100 |
Validated AA Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMTGT DISVPKPKYV ACPGVRIRNH EEFQYLDILA DVLSHGVLKP NRTGTDAYSK FGYQMRFDLS RSFPLLTTKK VALRSIIEEL LWFIKGSTNG NDLLAKNVRI WELNGRRDFL DKNGFTDREE HDLGPIYGFQ WRHFGAEYLD MHADYTGKGI DQLAEIINRI KTNPNDRRLI VCSWNVSDLK KMALPPCHCF FQFYVSDNKL SCMMHQRSCD LGLGVPFNIA SYSILTAMVA QVCGLGLGEF VHNLADAHIY VDHVDAVTTQ IARIPHPFPR LRLNPDIRNI EDFTIDDIVV EDY
Validated NT Sequence
sequence unavailable
Expected Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGNHEE FQYLDILADV LSHGVLKPNR TGTDAYSKFG YQMRFDLSRS FPLLTTKKVA LRSIIEELLW FIKGSTNGND LLAKNVRIWE LNGRRDFLDK NGFTDREEHD LGPIYGFQWR HFGAEYLDMH ADYTGKGIDQ LAEIINRIKT NPNDRRLIVC SWNVSDLKKM ALPPCHCFFQ FYVSDNKLSC MMHQRSCDLG LGVPFNIASY SILTAMVAQV CGLGLGEFVH NLADAHIYVD HVDAVTTQIA RIPHPFPRLR LNPDIRNIED FTIDDIVVED Y
Full NT Sequence (Expression Vector + Insert)
tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgttt acaatttcag gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc taaatacatt caaatatgta tccgctcatg aattaattct tagaaaaact catcgagcat caaatgaaac tgcaatttat tcatatcagg attatcaata ccatattttt gaaaaagccg tttctgtaat gaaggagaaa actcaccgag gcagttccat aggatggcaa gatcctggta tcggtctgcg attccgactc gtccaacatc aatacaacct attaatttcc cctcgtcaaa aataaggtta tcaagtgaga aatcaccatg agtgacgact gaatccggtg agaatggcaa aagtttatgc atttctttcc agacttgttc aacaggccag ccattacgct cgtcatcaaa atcactcgca tcaaccaaac cgttattcat tcgtgattgc gcctgagcga gacgaaatac gcgatcgctg ttaaaaggac aattacaaac aggaatcgaa tgcaaccggc gcaggaacac tgccagcgca tcaacaatat tttcacctga atcaggatat tcttctaata cctggaatgc tgttttcccg gggatcgcag tggtgagtaa ccatgcatca tcaggagtac ggataaaatg cttgatggtc ggaagaggca taaattccgt cagccagttt agtctgacca tctcatctgt aacatcattg gcaacgctac ctttgccatg tttcagaaac aactctggcg catcgggctt cccatacaat cgatagattg tcgcacctga ttgcccgaca ttatcgcgag cccatttata cccatataaa tcagcatcca tgttggaatt taatcgcggc ctagagcaag acgtttcccg ttgaatatgg ctcataacac cccttgtatt actgtttatg taagcagaca gttttattgt tcatgaccaa aatcccttaa cgtgagtttt cgttccactg agcgtcagac cccgtagaaa agatcaaagg atcttcttga gatccttttt ttctgcgcgt aatctgctgc ttgcaaacaa aaaaaccacc gctaccagcg gtggtttgtt tgccggatca agagctacca actctttttc cgaaggtaac tggcttcagc agagcgcaga taccaaatac tgtccttcta gtgtagccgt agttaggcca ccacttcaag aactctgtag caccgcctac atacctcgct ctgctaatcc tgttaccagt ggctgctgcc agtggcgata agtcgtgtct taccgggttg gactcaagac gatagttacc ggataaggcg cagcggtcgg gctgaacggg gggttcgtgc acacagccca gcttggagcg aacgacctac accgaactga gatacctaca gcgtgagcta tgagaaagcg ccacgcttcc cgaagggaga aaggcggaca ggtatccggt aagcggcagg gtcggaacag gagagcgcac gagggagctt ccagggggaa acgcctggta tctttatagt cctgtcgggt ttcgccacct ctgacttgag cgtcgatttt tgtgatgctc gtcagggggg cggagcctat ggaaaaacgc cagcaacgcg gcctttttac ggttcctggc cttttgctgg ccttttgctc acatgttctt tcctgcgtta tcccctgatt ctgtggataa ccgtattacc gcctttgagt gagctgatac cgctcgccgc agccgaacga ccgagcgcag cgagtcagtg agcgaggaag cggaagagcg cctgatgcgg tattttctcc ttacgcatct gtgcggtatt tcacaccgca tatatggtgc actctcagta caatctgctc tgatgccgca tagttaagcc agtatacact ccgctatcgc tacgtgactg ggtcatggct gcgccccgac acccgccaac acccgctgac gcgccctgac gggcttgtct gctcccggca tccgcttaca gacaagctgt gaccgtctcc gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt ctgcctgttc atccgcgtcc agctcgttga gtttctccag aagcgttaat gtctggcttc tgataaagcg ggccatgtta agggcggttt tttcctgttt ggtcactgat gcctccgtgt aagggggatt tctgttcatg ggggtaatga taccgatgaa acgagagagg atgctcacga tacgggttac tgatgatgaa catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta atacagatgt aggtgttcca cagggtagcc agcagcatcc tgcgatgcag atccggaaca taatggtgca gggcgctgac ttccgcgttt ccagacttta cgaaacacgg aaaccgaaga ccattcatgt tgttgctcag gtcgcagacg ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg tgattcattc tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag gagcacgatc atgcgcaccc gtggggccgc catgccggcg ataatggcct gcttctcgcc gaaacgtttg gtggcgggac cagtgacgaa ggcttgagcg agggcgtgca agattccgaa taccgcaagc gacaggccga tcatcgtcgc gctccagcga aagcggtcct cgccgaaaat gacccagagc gctgccggca cctgtcctac gagttgcatg ataaagaaga cagtcataag tgcggcgacg atagtcatgc cccgcgccca ccggaaggag ctgactgggt tgaaggctct caagggcatc ggtcgagatc ccggtgccta atgagtgagc taacttacat taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgccag ggtggttttt cttttcacca gtgagacggg caacagctga ttgcccttca ccgcctggcc ctgagagagt tgcagcaagc ggtccacgct ggtttgcccc agcaggcgaa aatcctgttt gatggtggtt aacggcggga tataacatga gctgtcttcg gtatcgtcgt atcccactac cgagatatcc gcaccaacgc gcagcccgga ctcggtaatg gcgcgcattg cgcccagcgc catctgatcg ttggcaacca gcatcgcagt gggaacgatg ccctcattca gcatttgcat ggtttgttga aaaccggaca tggcactcca gtcgccttcc cgttccgcta tcggctgaat ttgattgcga gtgagatatt tatgccagcc agccagacgc agacgcgccg agacagaact taatgggccc gctaacagcg cgatttgctg gtgacccaat gcgaccagat gctccacgcc cagtcgcgta ccgtcttcat gggagaaaat aatactgttg atgggtgtct ggtcagagac atcaagaaat aacgccggaa cattagtgca ggcagcttcc acagcaatgg catcctggtc atccagcgga tagttaatga tcagcccact gacgcgttgc gcgagaagat tgtgcaccgc cgctttacag gcttcgacgc cgcttcgttc taccatcgac accaccacgc tggcacccag ttgatcggcg cgagatttaa tcgccgcgac aatttgcgac ggcgcgtgca gggccagact ggaggtggca acgccaatca gcaacgactg tttgcccgcc agttgttgtg ccacgcggtt gggaatgtaa ttcagctccg ccatcgccgc ttccactttt tcccgcgttt tcgcagaaac gtggctggcc tggttcacca cgcgggaaac ggtctgataa gagacaccgg catactctgc gacatcgtat aacgttactg gtttcacatt caccaccctg aattgactct cttccgggcg ctatcatgcc ataccgcgaa aggttttgcg ccattcgatg gtgtccggga tctcgacgct ctcccttatg cgactcctgc attaggaagc agcccagtag taggttgagg ccgttgagca ccgccgccgc aaggaatggt gcatgcaagg agatggcgcc caacagtccc ccggccacgg ggcctgccac catacccacg ccgaaacaag cgctcatgag cccgaagtgg cgagcccgat cttccccatc ggtgatgtcg gcgatatagg cgccagcaac cgcacctgtg gcgccggtga tgccggccac gatgcgtccg gcgtagagga tcgagatctc gatcccgcga aattaatacg actcactata ggggaattgt gagcggataa caattcccct ctagaaataa ttttgtttaa ctttaagaag gagatatacc atgggccacc accaccacca ccatagcggc gaagtaaaac cggaagtgaa gccggagacc cacatcaacc tgaaggttag cgacggtagc agcgaaatct tcttcaagat taagaagacc accccgctgc gtcgcctgat ggaagcattc gctaaacgcc agggcaagga aatggattcc ctgcgctttc tgtacgacgg tatccgtatt caggcagacc agactccgga agatctggac atggaagata acgacattat cgaagcacac cgtgaacaaa tcggtggaaa ccacgaagaa ttccagtatc ttgacatctt agctgacgtg ctgtcccacg gtgtgctgaa gccgaaccgc accggcactg acgcttactc gaaatttggt taccagatgc gcttcgacct ttcccgtagc ttcccgctcc tgaccactaa gaaggttgcg ctccgctcta tcatcgagga actgctctgg tttatcaaag gttctaccaa cggtaacgat ctgttagcta agaatgttcg tatctgggaa ctgaacggtc gtcgtgactt tcttgacaag aatggtttca ctgatcgcga agagcacgac ctgggtccga tttacggctt ccagtggcgt cactttggtg ctgaatacct tgacatgcac gctgattaca ctggcaaagg tatcgatcag ttagctgaaa tcattaatcg catcaagact aacccgaacg atcgccgtct gatcgtttgc tcctggaacg tctctgatct caagaaaatg gcactgccgc cttgtcactg cttcttccag ttctatgtta gcgacaacaa actgagctgc atgatgcacc agcgtagctg cgatctgggc ctgggcgttc cgttcaatat cgcttcctac tccattctga ccgctatggt tgcccaagtt tgtggtcttg gtctgggtga attcgttcat aacctggctg atgcccacat ttacgtggac cacgttgacg ctgtgaccac ccagatagct cgtattcctc acccatttcc gcgcctgcgt cttaatccag acattcgtaa cattgaagac ttcaccatcg acgatatcgt cgtggaagat tactaaaagc ttgcggccgc actcgagcac caccaccacc accactgaga tccggctgct aacaaagccc gaaaggaagc tgagttggct gctgccaccg ctgagcaata actagcataa ccccttgggg cctctaaacg ggtcttgagg ggttttttgc tgaaaggagg aactatatcc ggat
Details for BaboA.01191.a.D13.PD00068
| PURIFICATION DATe: | 6/26/2009 |
| CONCENTRATION: | 27.2mg/ml |
| OBSERVED MW: | data unavailable |
| EXPRESSION LEVEL: | High Expression |
| PROTEIN PURIFICATION BUFFER: | 20mM Tris-HCl, pH 8.0, 200mM NaCl, 1% glycerol, 1mM TCEP |
| EXPRESSION HOST: | data unavailable |
| VIAL COUNT (approx.): | 1 |
| VIAL VOLUME: | 50µl |
| PERCENT IDENTITY: | 100 |
| PERCENT COVERAGE: | 61 |
Protocol Notes
notes unavailable
Validated AA Sequence
MRFDLSRSFP LLTTKKVALR SIIEELLWFI KGSTNGNDLL AKNVRIWELN GRRDFLDKNG FTDREEHDLG PIYGFQWRHF GAEYLDMHAD YTGKGIDQLA EIINRIKTNP NDRRLIVCSW NVSDLKKMAL PPCHCFFQFY VSDNKLSCMM HQRSCDLGLG VPFNIASYSI LTAMVAQVCG LGLGEFVHNL ADAHIYVDHV DAVTTQIARI PHPFPRLRLN PDIRNIEDFT IDDIVVEDYV SHPPIPMAMS A
Validated NT Sequence
ttcnggcttt gttagcagcc ggntctcngt ggtggtggtg gtggtgctcg agtgcggccg caagctttta tgcagacatg gccataggaa tcggtgggtg ggacacgtaa tcttccacga cgatatcgtc gatggtgaag tcttcaatgt tacgaatgtc tggattaaga cgcaggcgcg gaaatgggtg aggaatacga gctatctggg tggtcacagc gtcaacgtgg tccacgtaaa tgtgggcatc agccaggtta tgaacgaatt cacccagacc aagaccacaa acttgggcaa ccatagcggt cagaatggag taggaagcga tattgaacgg aacgcccagg cccagatcgc agctacgctg gtgcatcatg cagctcagtt tgttgtcgct aacatagaac tggaagaagc agtgacaagg cggcagtgcc attttcttga gatcagagac gttccaggag caaacgatca gacggcgatc gttcgggtta gtcttgatgc gattaatgat ttcagctaac tgatcgatac ctttgccagt gtaatcagcg tgcatgtcaa ggtattcagc accaaagtga cgccactgga agccgtaaat cggacccagg tcgtgctctt cgcgatcagt gaaaccattc ttgtcaagaa agtcacgacg accgttcagt tcccagatac gaacattctt agctaacaga tcgttaccgt tggtagaacc tttgataaac cagagcagtt cctcgatgat agagcggagc gcaaccttct tagtggtcag gagcgggaag ctacgggaaa ggtcgaagcg catctggtaa ccaaatttcg agtaagcgtc agtgccngtg cggttcggct tcagcacacc gtgggacagc acgtcagcta agatgtcnag atactggaat tcttcgtggt tnnngatacg aacgccnggn nggctacgta cttcggtttn nnaacagann ngtcggtgnc gntcattcca ccgattnnnn cangnnnnnc ntcgannatg tcntnntcnt nnntgtncag atnntnnnnn nnnngnctnn cnnantangn nnnccnnnnn nnaaagcnnn nnnntnnnnt tnnnnnnncn nnnnntnnnn nanngnnncn nnngnnnnnn nnnnngnnnn nnnnnnnnnn nnanancnnn nnnnnnnnnn gnnnnaannn nnnnnnnccg g
Expressed Protein Sequence
MGHHHHHHSG EVKPEVKPET HINLKVSDGS SEIFFKIKKT TPLRRLMEAF AKRQGKEMDS LRFLYDGIRI QADQTPEDLD MEDNDIIEAH REQIGGMTGT DISVPKPKYV ACPGVRIRNH EEFQYLDILA DVLSHGVLKP NRTGTDAYSK FGYQMRFDLS RSFPLLTTKK VALRSIIEEL LWFIKGSTNG NDLLAKNVRI WELNGRRDFL DKNGFTDREE HDLGPIYGFQ WRHFGAEYLD MHADYTGKGI DQLAEIINRI KTNPNDRRLI VCSWNVSDLK KMALPPCHCF FQFYVSDNKL SCMMHQRSCD LGLGVPFNIA SYSILTAMVA QVCGLGLGEF VHNLADAHIY VDHVDAVTTQ IARIPHPFPR LRLNPDIRNI EDFTIDDIVV EDYVSHPPIP MAMSA
Full NT Sequence (Expression Vector + Insert)
TGGCGAATGG GACGCGCCCT GTAGCGGCGC ATTAAGCGCG GCGGGTGTGG TGGTTACGCG CAGCGTGACC GCTACACTTG CCAGCGCCCT AGCGCCCGCT CCTTTCGCTT TCTTCCCTTC CTTTCTCGCC ACGTTCGCCG GCTTTCCCCG TCAAGCTCTA AATCGGGGGC TCCCTTTAGG GTTCCGATTT AGTGCTTTAC GGCACCTCGA CCCCAAAAAA CTTGATTAGG GTGATGGTTC ACGTAGTGGG CCATCGCCCT GATAGACGGT TTTTCGCCCT TTGACGTTGG AGTCCACGTT CTTTAATAGT GGACTCTTGT TCCAAACTGG AACAACACTC AACCCTATCT CGGTCTATTC TTTTGATTTA TAAGGGATTT TGCCGATTTC GGCCTATTGG TTAAAAAATG AGCTGATTTA ACAAAAATTT AACGCGAATT TTAACAAAAT ATTAACGTTT ACAATTTCAG GTGGCACTTT TCGGGGAAAT GTGCGCGGAA CCCCTATTTG TTTATTTTTC TAAATACATT CAAATATGTA TCCGCTCATG AATTAATTCT TAGAAAAACT CATCGAGCAT CAAATGAAAC TGCAATTTAT TCATATCAGG ATTATCAATA CCATATTTTT GAAAAAGCCG TTTCTGTAAT GAAGGAGAAA ACTCACCGAG GCAGTTCCAT AGGATGGCAA GATCCTGGTA TCGGTCTGCG ATTCCGACTC GTCCAACATC AATACAACCT ATTAATTTCC CCTCGTCAAA AATAAGGTTA TCAAGTGAGA AATCACCATG AGTGACGACT GAATCCGGTG AGAATGGCAA AAGTTTATGC ATTTCTTTCC AGACTTGTTC AACAGGCCAG CCATTACGCT CGTCATCAAA ATCACTCGCA TCAACCAAAC CGTTATTCAT TCGTGATTGC GCCTGAGCGA GACGAAATAC GCGATCGCTG TTAAAAGGAC AATTACAAAC AGGAATCGAA TGCAACCGGC GCAGGAACAC TGCCAGCGCA TCAACAATAT TTTCACCTGA ATCAGGATAT TCTTCTAATA CCTGGAATGC TGTTTTCCCG GGGATCGCAG TGGTGAGTAA CCATGCATCA TCAGGAGTAC GGATAAAATG CTTGATGGTC GGAAGAGGCA TAAATTCCGT CAGCCAGTTT AGTCTGACCA TCTCATCTGT AACATCATTG GCAACGCTAC CTTTGCCATG TTTCAGAAAC AACTCTGGCG CATCGGGCTT CCCATACAAT CGATAGATTG TCGCACCTGA TTGCCCGACA TTATCGCGAG CCCATTTATA CCCATATAAA TCAGCATCCA TGTTGGAATT TAATCGCGGC CTAGAGCAAG ACGTTTCCCG TTGAATATGG CTCATAACAC CCCTTGTATT ACTGTTTATG TAAGCAGACA GTTTTATTGT TCATGACCAA AATCCCTTAA CGTGAGTTTT CGTTCCACTG AGCGTCAGAC CCCGTAGAAA AGATCAAAGG ATCTTCTTGA GATCCTTTTT TTCTGCGCGT AATCTGCTGC TTGCAAACAA AAAAACCACC GCTACCAGCG GTGGTTTGTT TGCCGGATCA AGAGCTACCA ACTCTTTTTC CGAAGGTAAC TGGCTTCAGC AGAGCGCAGA TACCAAATAC TGTCCTTCTA GTGTAGCCGT AGTTAGGCCA CCACTTCAAG AACTCTGTAG CACCGCCTAC ATACCTCGCT CTGCTAATCC TGTTACCAGT GGCTGCTGCC AGTGGCGATA AGTCGTGTCT TACCGGGTTG GACTCAAGAC GATAGTTACC GGATAAGGCG CAGCGGTCGG GCTGAACGGG GGGTTCGTGC ACACAGCCCA GCTTGGAGCG AACGACCTAC ACCGAACTGA GATACCTACA GCGTGAGCTA TGAGAAAGCG CCACGCTTCC CGAAGGGAGA AAGGCGGACA GGTATCCGGT AAGCGGCAGG GTCGGAACAG GAGAGCGCAC GAGGGAGCTT CCAGGGGGAA ACGCCTGGTA TCTTTATAGT CCTGTCGGGT TTCGCCACCT CTGACTTGAG CGTCGATTTT TGTGATGCTC GTCAGGGGGG CGGAGCCTAT GGAAAAACGC CAGCAACGCG GCCTTTTTAC GGTTCCTGGC CTTTTGCTGG CCTTTTGCTC ACATGTTCTT TCCTGCGTTA TCCCCTGATT CTGTGGATAA CCGTATTACC GCCTTTGAGT GAGCTGATAC CGCTCGCCGC AGCCGAACGA CCGAGCGCAG CGAGTCAGTG AGCGAGGAAG CGGAAGAGCG CCTGATGCGG TATTTTCTCC TTACGCATCT GTGCGGTATT TCACACCGCA TATATGGTGC ACTCTCAGTA CAATCTGCTC TGATGCCGCA TAGTTAAGCC AGTATACACT CCGCTATCGC TACGTGACTG GGTCATGGCT GCGCCCCGAC ACCCGCCAAC ACCCGCTGAC GCGCCCTGAC GGGCTTGTCT GCTCCCGGCA TCCGCTTACA GACAAGCTGT GACCGTCTCC GGGAGCTGCA TGTGTCAGAG GTTTTCACCG TCATCACCGA AACGCGCGAG GCAGCTGCGG TAAAGCTCAT CAGCGTGGTC GTGAAGCGAT TCACAGATGT CTGCCTGTTC ATCCGCGTCC AGCTCGTTGA GTTTCTCCAG AAGCGTTAAT GTCTGGCTTC TGATAAAGCG GGCCATGTTA AGGGCGGTTT TTTCCTGTTT GGTCACTGAT GCCTCCGTGT AAGGGGGATT TCTGTTCATG GGGGTAATGA TACCGATGAA ACGAGAGAGG ATGCTCACGA TACGGGTTAC TGATGATGAA CATGCCCGGT TACTGGAACG TTGTGAGGGT AAACAACTGG CGGTATGGAT GCGGCGGGAC CAGAGAAAAA TCACTCAGGG TCAATGCCAG CGCTTCGTTA ATACAGATGT AGGTGTTCCA CAGGGTAGCC AGCAGCATCC TGCGATGCAG ATCCGGAACA TAATGGTGCA GGGCGCTGAC TTCCGCGTTT CCAGACTTTA CGAAACACGG AAACCGAAGA CCATTCATGT TGTTGCTCAG GTCGCAGACG TTTTGCAGCA GCAGTCGCTT CACGTTCGCT CGCGTATCGG TGATTCATTC TGCTAACCAG TAAGGCAACC CCGCCAGCCT AGCCGGGTCC TCAACGACAG GAGCACGATC ATGCGCACCC GTGGGGCCGC CATGCCGGCG ATAATGGCCT GCTTCTCGCC GAAACGTTTG GTGGCGGGAC CAGTGACGAA GGCTTGAGCG AGGGCGTGCA AGATTCCGAA TACCGCAAGC GACAGGCCGA TCATCGTCGC GCTCCAGCGA AAGCGGTCCT CGCCGAAAAT GACCCAGAGC GCTGCCGGCA CCTGTCCTAC GAGTTGCATG ATAAAGAAGA CAGTCATAAG TGCGGCGACG ATAGTCATGC CCCGCGCCCA CCGGAAGGAG CTGACTGGGT TGAAGGCTCT CAAGGGCATC GGTCGAGATC CCGGTGCCTA ATGAGTGAGC TAACTTACAT TAATTGCGTT GCGCTCACTG CCCGCTTTCC AGTCGGGAAA CCTGTCGTGC CAGCTGCATT AATGAATCGG CCAACGCGCG GGGAGAGGCG GTTTGCGTAT TGGGCGCCAG GGTGGTTTTT CTTTTCACCA GTGAGACGGG CAACAGCTGA TTGCCCTTCA CCGCCTGGCC CTGAGAGAGT TGCAGCAAGC GGTCCACGCT GGTTTGCCCC AGCAGGCGAA AATCCTGTTT GATGGTGGTT AACGGCGGGA TATAACATGA GCTGTCTTCG GTATCGTCGT ATCCCACTAC CGAGATATCC GCACCAACGC GCAGCCCGGA CTCGGTAATG GCGCGCATTG CGCCCAGCGC CATCTGATCG TTGGCAACCA GCATCGCAGT GGGAACGATG CCCTCATTCA GCATTTGCAT GGTTTGTTGA AAACCGGACA TGGCACTCCA GTCGCCTTCC CGTTCCGCTA TCGGCTGAAT TTGATTGCGA GTGAGATATT TATGCCAGCC AGCCAGACGC AGACGCGCCG AGACAGAACT TAATGGGCCC GCTAACAGCG CGATTTGCTG GTGACCCAAT GCGACCAGAT GCTCCACGCC CAGTCGCGTA CCGTCTTCAT GGGAGAAAAT AATACTGTTG ATGGGTGTCT GGTCAGAGAC ATCAAGAAAT AACGCCGGAA CATTAGTGCA GGCAGCTTCC ACAGCAATGG CATCCTGGTC ATCCAGCGGA TAGTTAATGA TCAGCCCACT GACGCGTTGC GCGAGAAGAT TGTGCACCGC CGCTTTACAG GCTTCGACGC CGCTTCGTTC TACCATCGAC ACCACCACGC TGGCACCCAG TTGATCGGCG CGAGATTTAA TCGCCGCGAC AATTTGCGAC GGCGCGTGCA GGGCCAGACT GGAGGTGGCA ACGCCAATCA GCAACGACTG TTTGCCCGCC AGTTGTTGTG CCACGCGGTT GGGAATGTAA TTCAGCTCCG CCATCGCCGC TTCCACTTTT TCCCGCGTTT TCGCAGAAAC GTGGCTGGCC TGGTTCACCA CGCGGGAAAC GGTCTGATAA GAGACACCGG CATACTCTGC GACATCGTAT AACGTTACTG GTTTCACATT CACCACCCTG AATTGACTCT CTTCCGGGCG CTATCATGCC ATACCGCGAA AGGTTTTGCG CCATTCGATG GTGTCCGGGA TCTCGACGCT CTCCCTTATG CGACTCCTGC ATTAGGAAGC AGCCCAGTAG TAGGTTGAGG CCGTTGAGCA CCGCCGCCGC AAGGAATGGT GCATGCAAGG AGATGGCGCC CAACAGTCCC CCGGCCACGG GGCCTGCCAC CATACCCACG CCGAAACAAG CGCTCATGAG CCCGAAGTGG CGAGCCCGAT CTTCCCCATC GGTGATGTCG GCGATATAGG CGCCAGCAAC CGCACCTGTG GCGCCGGTGA TGCCGGCCAC GATGCGTCCG GCGTAGAGGA TCGAGATCTC GATCCCGCGA AATTAATACG ACTCACTATA GGGGAATTGT GAGCGGATAA CAATTCCCCT CTAGAAATAA TTTTGTTTAA CTTTAAGAAG GAGATATACC ATGGGCCACC ACCACCACCA CCATAGCGGC GAAGTAAAAC CGGAAGTGAA GCCGGAGACC CACATCAACC TGAAGGTTAG CGACGGTAGC AGCGAAATCT TCTTCAAGAT TAAGAAGACC ACCCCGCTGC GTCGCCTGAT GGAAGCATTC GCTAAACGCC AGGGCAAGGA AATGGATTCC CTGCGCTTTC TGTACGACGG TATCCGTATT CAGGCAGACC AGACTCCGGA AGATCTGGAC ATGGAAGATA ACGACATTAT CGAAGCACAC CGTGAACAAA TCGGTGGAAT GACCGGCACC GACATCTCTG TTCCTAAACC GAAGTACGTA GCCTGCCCTG GCGTTCGTAT CCGTAACCAC GAAGAATTCC AGTATCTTGA CATCTTAGCT GACGTGCTGT CCCACGGTGT GCTGAAGCCG AACCGCACCG GCACTGACGC TTACTCGAAA TTTGGTTACC AGATGCGCTT CGACCTTTCC CGTAGCTTCC CGCTCCTGAC CACTAAGAAG GTTGCGCTCC GCTCTATCAT CGAGGAACTG CTCTGGTTTA TCAAAGGTTC TACCAACGGT AACGATCTGT TAGCTAAGAA TGTTCGTATC TGGGAACTGA ACGGTCGTCG TGACTTTCTT GACAAGAATG GTTTCACTGA TCGCGAAGAG CACGACCTGG GTCCGATTTA CGGCTTCCAG TGGCGTCACT TTGGTGCTGA ATACCTTGAC ATGCACGCTG ATTACACTGG CAAAGGTATC GATCAGTTAG CTGAAATCAT TAATCGCATC AAGACTAACC CGAACGATCG CCGTCTGATC GTTTGCTCCT GGAACGTCTC TGATCTCAAG AAAATGGCAC TGCCGCCTTG TCACTGCTTC TTCCAGTTCT ATGTTAGCGA CAACAAACTG AGCTGCATGA TGCACCAGCG TAGCTGCGAT CTGGGCCTGG GCGTTCCGTT CAATATCGCT TCCTACTCCA TTCTGACCGC TATGGTTGCC CAAGTTTGTG GTCTTGGTCT GGGTGAATTC GTTCATAACC TGGCTGATGC CCACATTTAC GTGGACCACG TTGACGCTGT GACCACCCAG ATAGCTCGTA TTCCTCACCC ATTTCCGCGC CTGCGTCTTA ATCCAGACAT TCGTAACATT GAAGACTTCA CCATCGACGA TATCGTCGTG GAAGATTACG TGTCCCACCC ACCGATTCCT ATGGCCATGT CTGCATAAAA GCTTGCGGCC GCACTCGAGC ACCACCACCA CCACCACTGA GATCCGGCTG CTAACAAAGC CCGAAAGGAA GCTGAGTTGG CTGCTGCCAC CGCTGAGCAA TAACTAGCAT AACCCCTTGG GGCCTCTAAA CGGGTCTTGA GGGGTTTTTT GCTGAAAGGA GGAACTATAT CCGGAT